Calcium-sensing receptor database (CASRdb)
A Publicly Accessible Comprehensive Database for Calcium-sensing Receptor (CASR) Variants Associated with Disorders of Calcium Metabolism
Introduction
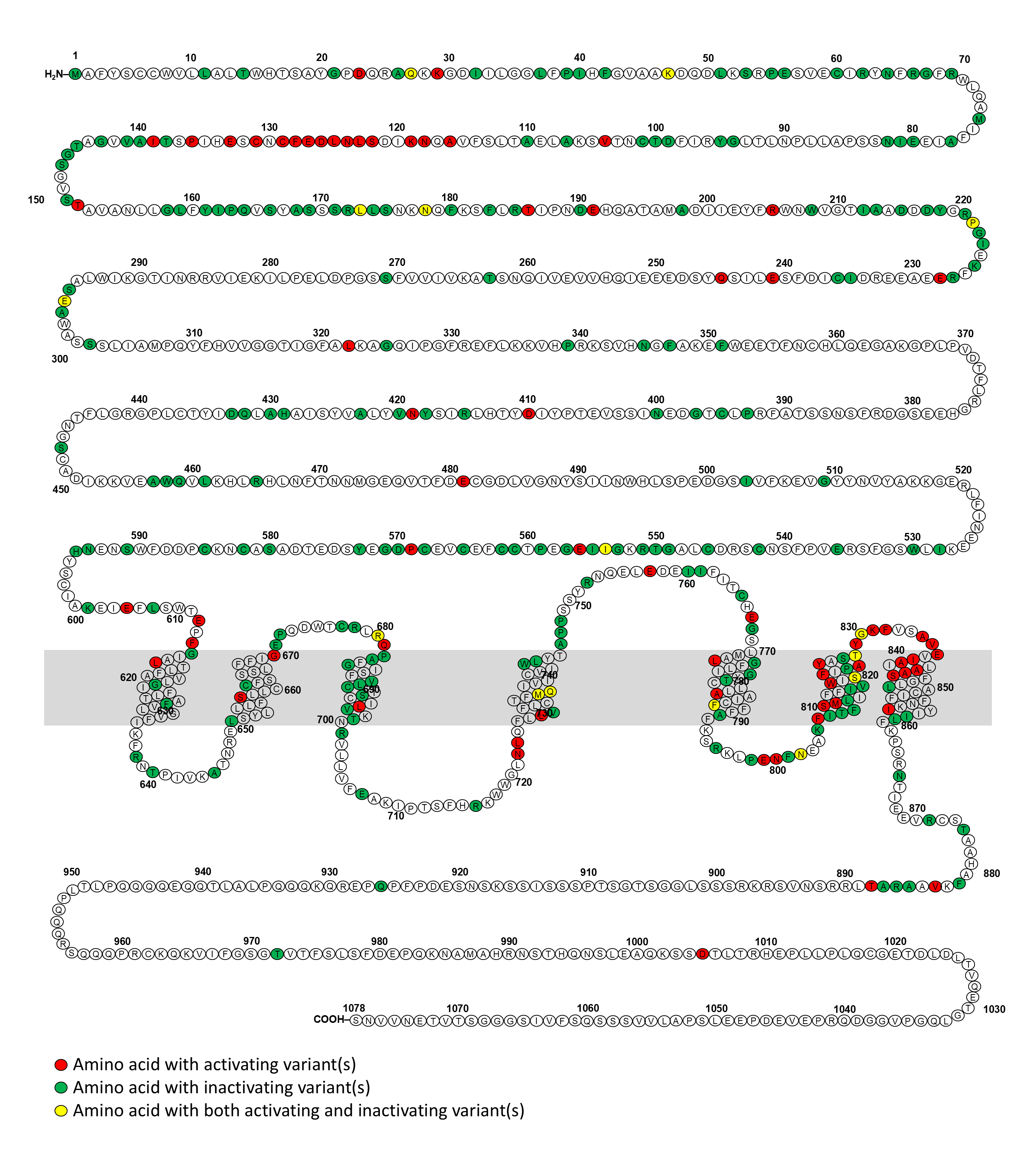
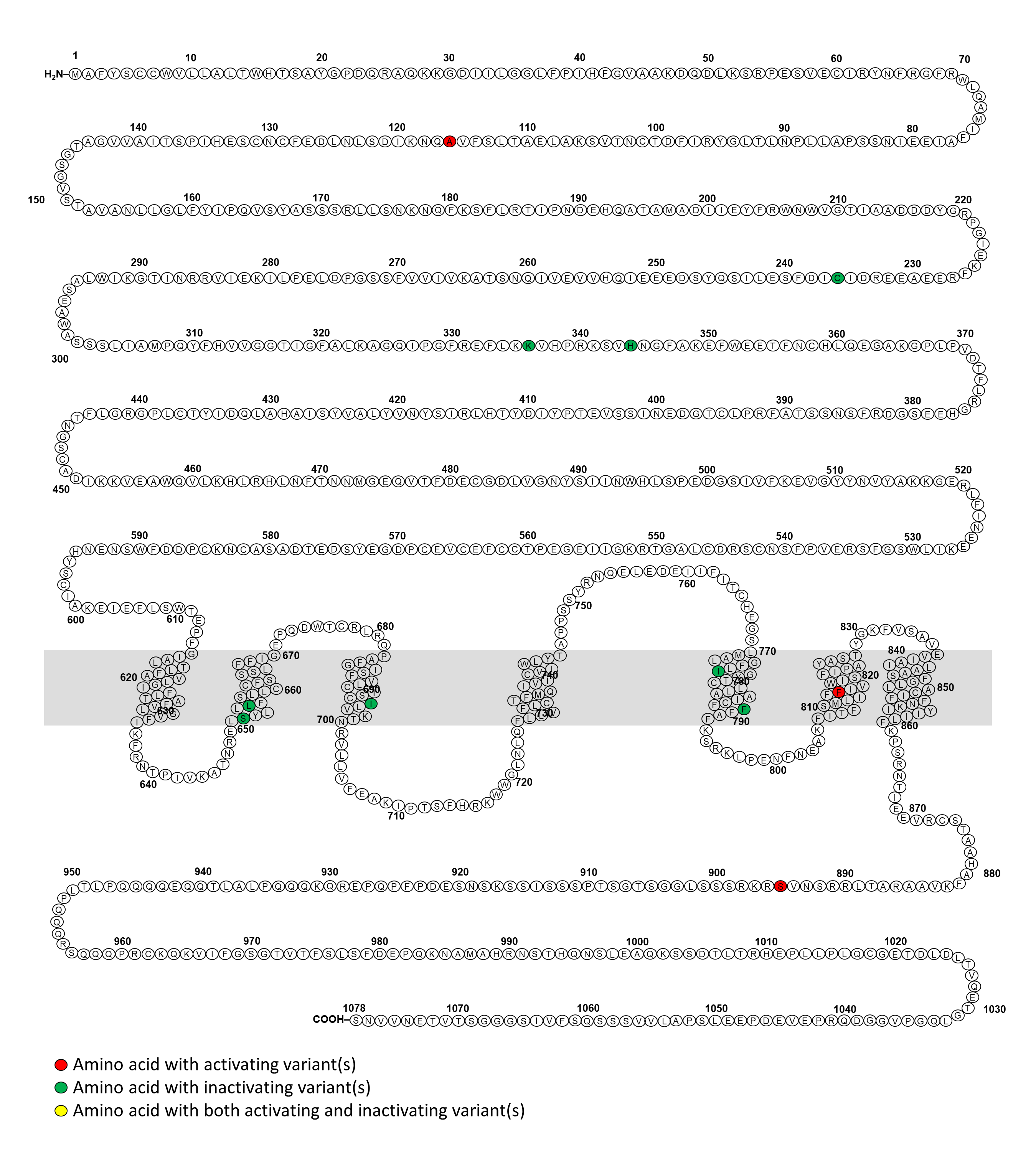
The calcium-sensing receptor (CaSR) is a 1,078 amino acid G protein-coupled receptor. It is highly expressed in the parathyroid glands and kidney where is responsible for regulating calcium homeostasis (1). Inactivating mutations in the CASR gene cause familial hypocalciuric hypercalcemia (FHH) type I and neonatal severe hyperparathyroidism, while activating mutations of the CASR result in autosomal dominant hypocalcemia (ADH) type I and autosomal dominant hypocalcemia with Bartter syndrome (previously known as Bartter syndrome type V) (1). Genetic testing of CASR is crucial for confirming the clinical diagnosis of these conditions. As the McGill’s CASRdb (2) is no longer available, there is currently a lack of a comprehensive database of variants of the CASR gene and their disease associations.
We created this website with the aim to present a publicly accessible comprehensive database with interactive visualization of the disease-causing variants of the CASR gene based on available literature and genomic databases. We hope that this CASRdb will be a comprehensive resource aiding clinicians in interpreting CASR variants and assisting researchers in variant curation and structure-function analyses.
References
1. Hannan FM, Kallay E, Chang W, Brandi ML, Thakker RV. The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases. Nat Rev Endocrinol. 2018;15(1):33-51.
2. Pidasheva S, D’Souza-Li L, Canaff L, Cole DE, Hendy GN. CASRdb: calcium-sensing receptor locus-specific database for mutations causing familial (benign) hypocalciuric hypercalcemia, neonatal severe hyperparathyroidism, and autosomal dominant hypocalcemia. Hum Mutat. 2004;24(2):107-11.
Methodology
We conducted a systematic review of the Embase and Pubmed databases from inception to March 2023, using a search strategy associated with “Calcium-sensing receptor”. Additionally, we identified supporting references for pathogenic or likely pathogenic variants reported in the ClinVar and LOVD databases. We compiled a comprehensive list of CASR variants associated with disorders of calcium metabolism. Benign or likely benign variants were excluded. Variants of uncertain significance in the ClinVar and LOVD databases were included only if they were reported in the literature to be associated with disorders of the CaSR. This list has been made available on this website equipped with a search engine and links to the respective references.
Gene Structure
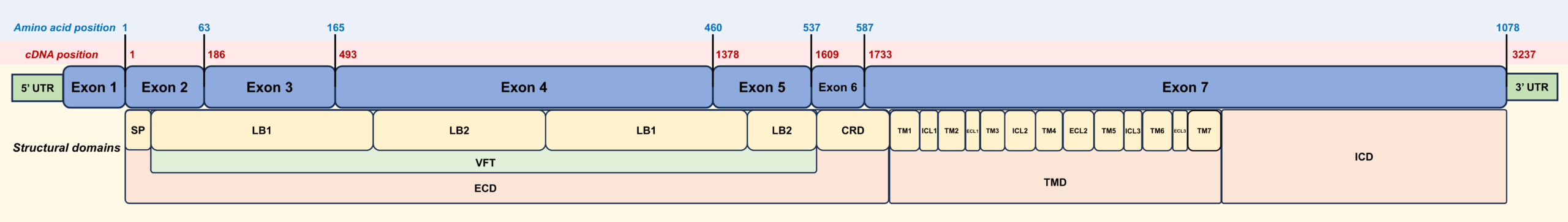
Abbreviations: SP: Signal Peptide; VFT: Venus Flytrap Domain; LB1: Lobe 1; LB2: Lobe 2; CRD: Cysteine-rich Domain; TMD: Transmembrane Domain; ECL1: Extracellular Loop 1; ECL2: Extracellular Loop 2; ECL3: Extracellular Loop 3; ICL1: Intracellular Loop 1; ICL2: Intracellular Loop 2; ICL3: Intracellular Loop 3; ICD: Intracellular Domain
Protein Structure
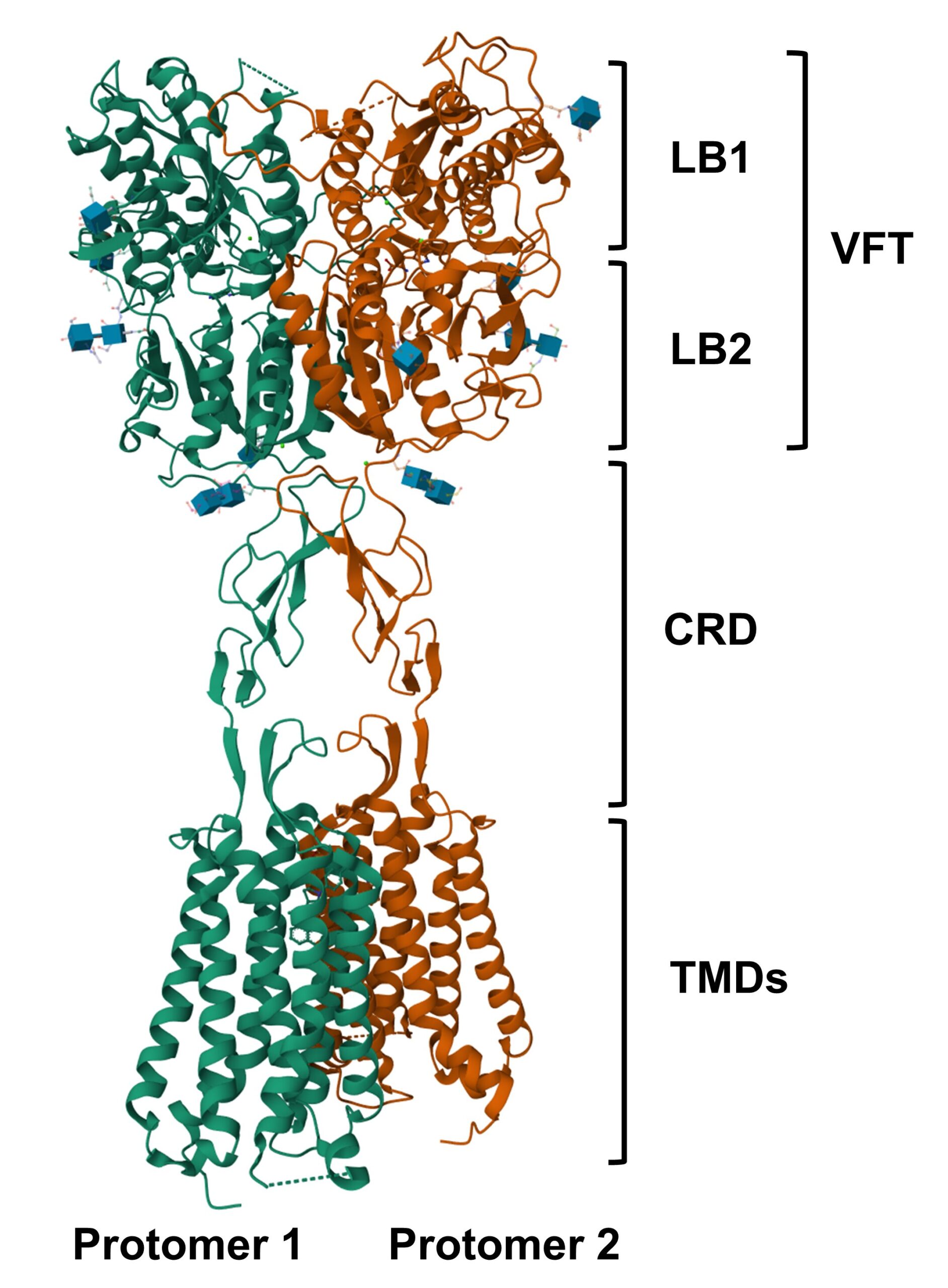
Image retrieved from the RCSB Protein Data Bank
Structure of calcium-sensing receptor in complex with evocalcet, a calcimimetics
and derivative of cinacalcet. Protomer 1 in green, Protomer 2 in orange.
Explore more in 3D: Structure | Sequence Annotations | Ligand Interaction
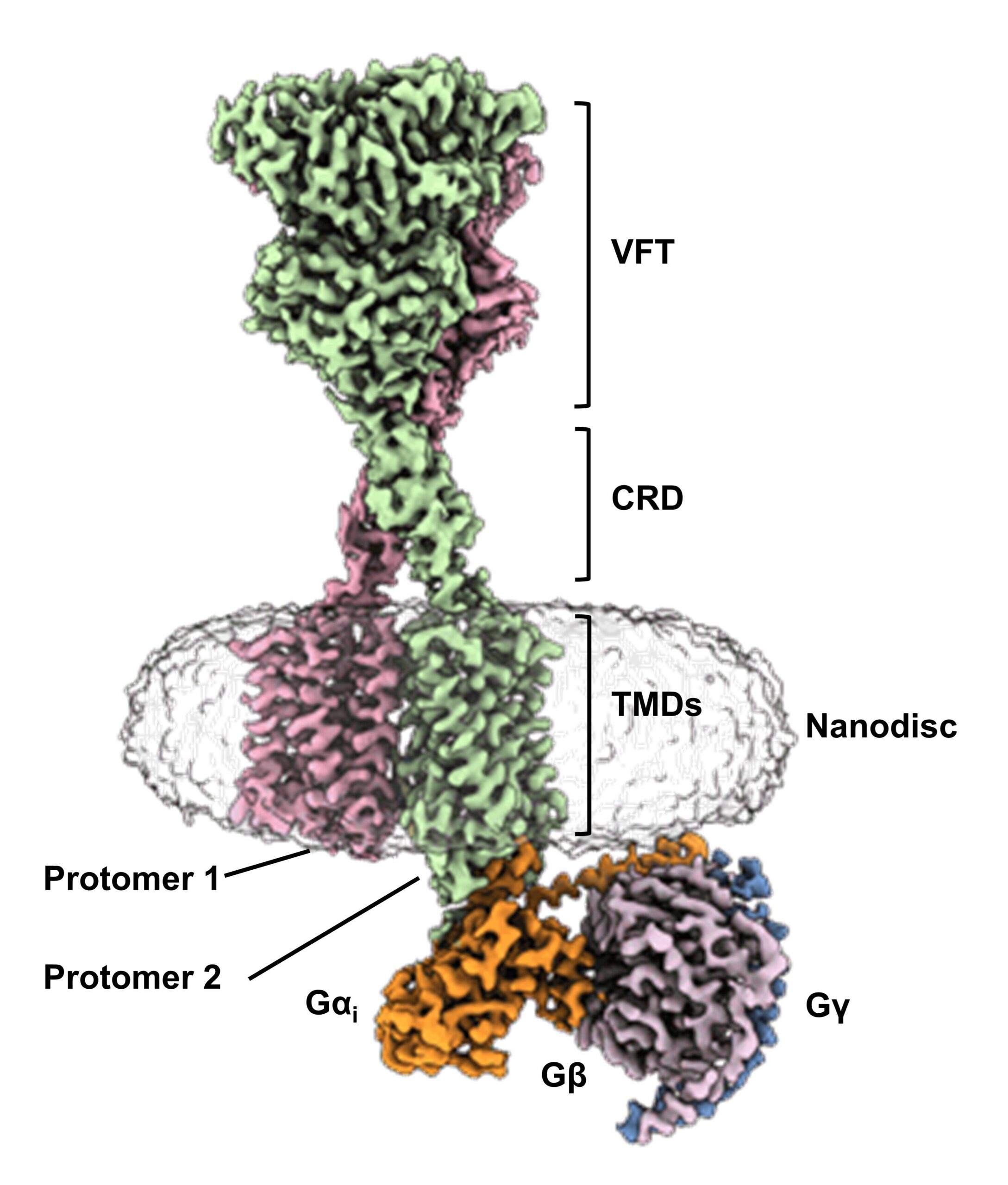
Image retrieved from the Electron Microscopy Data Bank
Cryo-EM structure of human calcium-sensing receptor-Gi complex in lipid nanodiscs
Protomer 1 in pink, Protomer 2 in green, Gαi in orange, Gβ in light purple, Gγ in blue.
View 3D structure
Abbreviations: LB1: Lobe 1; LB2: Lobe 2; VFT: Venus Flytrap Domain; CRD: Cysteine-rich Domain; TMDs: Transmembrane Domains
CASRdb
Scroll horizontally to view the remaining columns of the table.
Search by variant details to filter the table below (e.g., c.1A>G, p.Met1Val).
| cDNA Position | Variant | Amino acid change | Amino acid change | Variant effect | Clinical phenotype | Genomic co-ordinate | rs number | Variant type | Effect type | Location (gene) | Location (protein) | Pathogenicity reported in ClinVar | Article first author | Year | PMID | DOI/link | Article type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.1A>G | M1V | p.Met1Val | Inactivating | FHH1 | 122254190 | NA | SNV | Missense | Exon 2 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1321411/?oq=%22NM_000388.4(CASR):c.1A%3EG%20(p.Met1Val)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1A%3EG%20(p.Met1Val)#id_second | Clinvar |
| 2 | c.2T>G | M1R | p.Met1Arg | Inactivating | FHH1 (heterozygous) | 122254191 | NA | SNV | Missense | Exon 2 | ECD | NA | de Andrade | 2006 | 17121537 | 10.1111/j.1365-2265.2006.02640.x | Journal article |
| 2 | c.2T>G | M1R | p.Met1Arg | Inactivating | NSHPT (homozygous) | 122254191 | NA | SNV | Missense | Exon 2 | ECD | NA | de Andrade | 2006 | 17121537 | 10.1111/j.1365-2265.2006.02640.x | Journal article |
| 19 | c.19_20insT | C7fs*41 | p.Cys7fs*41 | Inactivating | FHH1 (heterozygous) | 122254208 | NA | Small insertion | Frameshift | Exon 2 | ECD | NA | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 19 | c.19_20insT | C7fs*41 | p.Cys7fs*41 | Inactivating | FHH1 | 122254208 | NA | Small insertion | Frameshift | Exon 2 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 23 | c.23G>A | W8* | p.Trp8* | Inactivating | FHH1 | 122254212 | NA | SNV | Nonsense | Exon 2 | ECD | Likely pathogenic | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 32 | c.32T>C | L11S | p.Leu11Ser | Inactivating | FHH1 | 122254221 | rs200673016 | SNV | Missense | Exon 2 | ECD | VUS | Pidasheva | 2005 | 15879434 | 10.1093/hmg/ddi176 | Journal article |
| 38 | c.38T>C | L13P | p.Leu13Pro | Inactivating | FHH1 | 122254227 | rs104893717 | SNV | Missense | Exon 2 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 38 | c.38T>C | L13P | p.Leu13Pro | Inactivating | FHH1 | 122254227 | rs104893717 | SNV | Missense | Exon 2 | ECD | Pathogenic | Miyashiro | 2004 | 15579740 | 10.1210/jc.2004-1046 | Journal article |
| 38 | c.38T>C | L13P | p.Leu13Pro | Inactivating | FHH1 | 122254227 | rs104893717 | SNV | Missense | Exon 2 | ECD | Pathogenic | Pidasheva | 2005 | 15879434 | 10.1093/hmg/ddi176 | Journal article |
| 61 | c.61G>A | G21R | p.Gly21Arg | Inactivating | FHH1 (heterozygous) | 122254250 | rs1064794290 | SNV | Missense | Exon 2 | ECD | VUS | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 69 | c.69C>A | D23E | p.Asp23Glu | Activating | ADH1 | 122254258 | NA | SNV | Missense | Exon 2 | ECD | NA | Dershem | 2020 | 32386559 | 10.1016/j.ajhg.2020.04.006 | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | FHH1 (heterozygous) | 122254262 | rs201633414 | SNV | Nonsense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | FHH1 (heterozygous) | 122254262 | rs201633414 | SNV | Missense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | Frank-Raue | 2011 | 21521328 | 10.1111/j.1365-2265.2011.04059.x | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | FHH1 (heterozygous) | 122254262 | rs201633414 | SNV | Missense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | FHH1 (heterozygous) | 122254262 | rs201633414 | SNV | Nonsense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | NSHPT (homozygous) | 122254262 | rs201633414 | SNV | Nonsense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 73 | c.73C>T | R25* | p.Arg25* | Inactivating | FHH1 (heterozygous) | 122254262 | rs201633414 | SNV | Nonsense | Exon 2 | ECD | VUS/Likely pathogenic/Pathogenic | Ward | 2006 | 16649980 | 10.1111/j.1365-2265.2006.02512.x | Journal article |
| 76 | c.76G>T | A26S | p.Ala26Ser | Inactivating | FHH1 (heterozygous) | 122254265 | NA | SNV | Missense | Exon 2 | ECD | NA | Salas | 2017 | NA | 10.1016/j.labcli.2017.03.001 | Journal article |
| 79 | c.79C>G | Q27E | p.Gln27Glu | Activating | ADH1 | 122254268 | NA | SNV | Missense | Exon 2 | ECD | NA | Chapman | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 79 | c.79C>G | Q27E | p.Gln27Glu | Activating | ADH1 | 122254268 | NA | SNV | Missense | Exon 2 | ECD | NA | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 80 | c.80A>G | Q27R | p.Gln27Arg | Inactivating | FHH1 (homozygous) | 122254269 | NA | SNV | Missense | Exon 2 | ECD | NA | Chikatsu | 1999 | 10468915 | 10.1046/j.1365-2265.1999.00729.x | Journal article |
| 80 | c.80A>C | Q27P | p.Gln27Pro | Inactivating | FHH1 | 122254269 | NA | SNV | Missense | Exon 2 | ECD | NA | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 80 | c.80A>G | Q27R | p.Gln27Arg | Inactivating | FHH1 | 122254269 | NA | SNV | Missense | Exon 2 | ECD | NA | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 85 | c.85A>G | K29E | p.Lys29Glu | Activating | ADH1 | 122254274 | rs397514729 | SNV | Missense | Exon 2 | ECD | Pathogenic | Hu | 2004 | 15005845 | 10.1359/JBMR.040106 | Journal article |
| 85 | c.85A>G | K29E | p.Lys29Glu | Activating | ADH1 | 122254274 | rs397514729 | SNV | Missense | Exon 2 | ECD | Pathogenic | Letz | 2014 | 25506941 | 10.1371/journal.pone.0115178 | Journal article |
| 85 | c.85A>G | K29E | p.Lys29Glu | Activating | ADH1 with Bartter syndrome | 122254274 | rs397514729 | SNV | Missense | Exon 2 | ECD | Pathogenic | Vezzoli | 2006 | 17048213 | NA | Journal article |
| 91 | c.91dup | D31fs* | p.Asp31fs* | Inactivating | FHH1 | 122254275 | NA | Small insertion | Frameshift | Exon 2 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1217243/?oq=%22NM_000388.4(CASR):c.91dup%20(p.Asp31fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.91dup%20(p.Asp31fs)#id_second | Clinvar |
| 94 | c.94A>G | I32V | p.Ile32Val | Inactivating | FHH1 (heterozygous) | 122254283 | NA | SNV | Missense | Exon 2 | ECD | VUS | Szalat | 2015 | 25091521 | 10.1007/s12020-014-0370-3 | Journal article |
| 94 | c.94A>G | I32V | p.Ile32Val | Inactivating | FHH1 (heterozygous) | 122254283 | NA | SNV | Missense | Exon 2 | ECD | VUS | Szalat | 2017 | 28176280 | 10.1007/s12020-017-1241-5 | Journal article |
| 108 | c.108_109insC | L37A*fs11 | p.Leu37Alafs*11 | Inactivating | FHH1 (heterozygous) | 122254297 | NA | Small insertion | Frameshift | Exon 2 | ECD | NA | Gurtunca | 2012 | NA | Gurtunca N, Yesilkaya E, Witchel S. A novel mutation in familial hypocalciuric hypercalcemia presenting in early infancy with irritability and mild hypercalcemia. Endocrine Reviews. 2012;33(3). | Conference abstract |
| 108 | c.108del | L37fs* | p.Leu37fs* | Inactivating | FHH1 | 122254292 | NA | Small deletion | Frameshift | Exon 2 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1321439/?oq=%22NM_000388.4(CASR):c.108del%20(p.Leu37fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.108del%20(p.Leu37fs)#id_second | Clinvar |
| 108 | c.108dup | L37fs* | p.Leu37fs* | NA | NA | 122254291 | rs886041823 | Small insertion | Frameshift | Exon 2 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/280657/?oq=%22NM_000388.4(CASR):c.108dup%20(p.Leu37fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.108dup%20(p.Leu37fs)#id_second | Clinvar |
| 110 | c.110T>C | L37P | p.Leu37Pro | Inactivating | FHH1 | 122254299 | NA | SNV | Missense | Exon 2 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 112 | c.112_113ins | F38fs* | p.Phe38fs* | NA | NA | 122254301 | NA | Small insertion | Frameshift | Exon 2 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1460173/?oq=%22NM_000388.4(CASR):c.112_113insC%20(p.Phe38fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.112_113insC%20(p.Phe38fs)#id_second | Clinvar |
| 115 | c.115C>G | P39A | p.Pro39Ala | Inactivating | NSHPT (homozygous) | 122254304 | rs121909262 | SNV | Missense | Exon 2 | ECD | Pathogenic | Abdullayev | 2020 | 31931451 | 10.1016/j.ijscr.2019.12.024 | Journal article |
| 115 | c.115C>G | P39A | p.Pro39Ala | Inactivating | FHH1 (homozygous) | 122254304 | rs121909262 | SNV | Missense | Exon 2 | ECD | Pathogenic | Chikatsu | 1999 | 10468915 | 10.1046/j.1365-2265.1999.00729.x | Journal article |
| 115 | c.115C>G | P39A | p.Pro39Ala | Inactivating | FHH1 | 122254304 | rs121909262 | SNV | Missense | Exon 2 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 118 | c.118A>T | I40F | p.Ile40Phe | Inactivating | FHH1 | 122254307 | NA | SNV | Missense | Exon 2 | ECD | NA | Vargas-Poussou | 2002 | 12191970 | 10.1097/01.asn.0000025781.16723.68 | Journal article |
| 122 | c.122A>G | H41R | p.His41Arg | Inactivating | FHH1 (heterozygous) | 122254311 | NA | SNV | Missense | Exon 2 | ECD | NA | Courtney | 2022 | 36536367 | 10.1186/s12902-022-01231-z | Journal article |
| 125 | c.125T>C | F42S | p.Phe42Ser | Inactivating | FHH1 | 122254314 | rs1553765909 | SNV | Missense | Exon 2 | ECD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 139 | c.139A>G | K47E | p.Lys47Glu | Inactivating | FHH1 | 122254328 | NA | SNV | Missense | Exon 2 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1685601/?oq=%22NM_000388.4(CASR):c.139A%3EG%20(p.Lys47Glu)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.139A%3EG%20(p.Lys47Glu)#id_second | Clinvar |
| 141 | c.141A>C | K47N | p.Lys47Asn | Activating | ADH1 | 122254330 | rs104893702 | SNV | Missense | Exon 2 | ECD | Pathogenic | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 141 | c.141A>C | K47N | p.Lys47Asn | Activating | ADH1 | 122254330 | rs104893702 | SNV | Missense | Exon 2 | ECD | Pathogenic | Okazaki | 1999 | 9920108 | 10.1210/jcem.84.1.5385 | Journal article |
| 152 | c.152T>G | L51R | p.Leu51Arg | Inactivating | FHH1 | 122254341 | NA | SNV | Missense | Exon 2 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 157 | c.157T>C | S53P | p.Ser53Pro | Inactivating | FHH1 (heterozygous) | 122254346 | NA | SNV | Missense | Exon 2 | ECD | Pathogenic | Alix | 2014 | 25444087 | 10.1016/j.jbspin.2014.08.007 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 164 | c.164C>T | P55R | p.Pro55Arg | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | López | 2011 | 21697018 | 10.1016/j.endonu.2011.04.004 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Quarde | 2021 | NA | 10.1016/j.jecr.2021.100093 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Speer | 2003 | 14714270 | 10.1055/s-2003-44708 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 (heterozygous) | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Sumida | 2022 | 35733207 | 10.1186/s12902-022-01077-5 | Journal article |
| 164 | c.164C>T | P55L | p.Pro55Leu | Inactivating | FHH1 | 122254353 | rs886041154 | SNV | Missense | Exon 2 | ECD | Pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 166 | c.166del | E56fs* | p.Glu56fs* | Inactivating | FHH1 | 122254354 | rs193922424 | Small deletion | Frameshift | Exon 2 | ECD | Pathogenic/Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35779/?oq=%22NM_000388.4(CASR):c.166del%20(p.Glu56fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.166del%20(p.Glu56fs)#id_second | Clinvar |
| 166 | c.166G>T | E56* | p.Glu56* | NA | NA | 122254355 | rs1358793834 | SNV | Nonsense | Exon 2 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/838073/?oq=%22NM_000388.4(CASR):c.166G%3ET%20(p.Glu56Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.166G%3ET%20(p.Glu56Ter)#id_first | Clinvar |
| 168 | c.168G>C | E56D | p.Glu56Asp | Inactivating | FHH1 (heterozygous) | 122254357 | NA | SNV | Missense | Exon 2 | ECD | NA | Park | 2022 | 35586626 | 10.3389/fendo.2022.853171 | Journal article |
| 178 | c.178T>C | C60R | p.Cys60Arg | Inactivating | FHH1 (heterozygous) | 122254367 | NA | SNV | Missense | Exon 2 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 178 | c.178T>G | C60G | p.Cys60Gly | Inactivating | FHH1 (homozygous) | 122254367 | NA | SNV | Missense | Exon 2 | ECD | NA | Li | 2021 | 34397587 | 10.1097/CM9.0000000000001568 | Journal article |
| 179 | c.179G>T | C60F | p.Cys60Phe | Inactivating | FHH1 (heterozygous) | 122254368 | rs772906030 | SNV | Missense | Exon 2 | ECD | VUS | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 179 | c.179G>A | C60Y | p.Cys60Tyr | Inactivating | FHH1 (heterozygous) | 122254368 | NA | SNV | Missense | Exon 2 | ECD | NA | Dong | 2020 | NA | 10.3760/cma.j.cn511374-20191118-00587 | Journal article |
| 179 | c.179G>A | C60Y | p.Cys60Tyr | Inactivating | NSHPT (compound heterozygous) | 122254368 | NA | SNV | Missense | Exon 2 | ECD | NA | Dong | 2020 | NA | 10.3760/cma.j.cn511374-20191118-00587 | Journal article |
| 179 | c.179G>T | C60F | p.Cys60Phe | Inactivating | NSHPT (compound heterozygous) | 122254368 | rs772906030 | SNV | Missense | Exon 2 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 185 | c.185G>T | R62M | p.Arg62Met | Inactivating | FHH1 | 122254374 | rs121909265 | SNV | Missense | Exon 2 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 185 | c.185G>T | R62M | p.Arg62Met | Inactivating | FHH1 (heterozygous) | 122254374 | rs121909265 | SNV | Missense | Exon 2 | ECD | Pathogenic | Chou | 1995 | 7726161 | NA | Journal article |
| 185 | c.185+1G>C | NA | NA | Inactivating | FHH1 (heterozygous) | 122254375 | NA | SNV | Splice donor | Exon 2-Intron 2 splice site | ECD | NA | Cetani | 2009 | 19073830 | 10.1530/EJE-08-0798 | Journal article |
| 185 | c.185+2T>C | NA | NA | Inactivating | FHH1 | 122254376 | NA | SNV | Splice donor | Exon 2-Intron 2 splice site | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 186 | c186-1G>T | R62_164delfs*94 | p.Arg62_164delfs*94 | Inactivating | FHH1 (heterozygous) | 122257080 | rs797044441 | SNV | Splice acceptor/frameshift | Intron 2-Exon 3 splice site | ECD | Pathogenic | D'Souza-Li | 2001 | 11668634 | 10.1002/humu.1212 | Journal article |
| 186 | c.186del | R62Sfs*13 | p.Arg62Serfs*13 | Inactivating | FHH1 | 122257081 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 190 | c.190A>G | N64D | p.Asn64Asp | Inactivating | FHH1 | 122257085 | NA | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1698732/?oq=%22NM_000388.4(CASR):c.190A%3EG%20(p.Asn64Asp)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.190A%3EG%20(p.Asn64Asp)#id_second | Clinvar |
| 196 | c.196C>T | R66C | p.Arg66Cys | Inactivating | FHH1 (heterozygous) | 122257091 | rs121909266 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Chou | 1995 | 7726161 | NA | Journal article |
| 196 | c.196C>T | R66C | p.Arg66Cys | Inactivating | FHH1 | 122257091 | rs121909266 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Pidasheva | 2006 | 16740594 | 10.1093/hmg/ddl145 | Journal article |
| 197 | c.197G>A | R66H | p.Arg66His | Inactivating | FHH1 (heterozygous) | 122257092 | rs1276839362 | SNV | Missense | Exon 3 | ECD | Pathogenic/Likely pathogenic | Misof | 2012 | NA | 10.1016/j.bone.2012.08.049 | Conference abstract |
| 197 | c.197G>A | R66H | p.Arg66His | Inactivating | FHH1 | 122257092 | rs1276839362 | SNV | Missense | Exon 3 | ECD | Pathogenic/Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 197 | c.197G>A | R66H | p.Arg66His | Inactivating | FHH1 | 122257092 | rs1276839362 | SNV | Missense | Exon 3 | ECD | Pathogenic/Likely pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 197 | c.197G>A | R66H | p.Arg66His | Inactivating | FHH1 | 122257092 | rs1276839362 | SNV | Missense | Exon 3 | ECD | Pathogenic/Likely pathogenic | Pidasheva | 2006 | 16740594 | 10.1093/hmg/ddl145 | Journal article |
| 199 | c.199delinsTTCG | G67fs* | p.Gly67fs* | NA | NA | 122257094 | rs1559956508 | Small insertion/deletion | Frameshift | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/574086/?oq=%22NM_000388.4(CASR):c.199delinsTTCGCT%20(p.Gly67fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.199delinsTTCGCT%20(p.Gly67fs)#id_second | Clinvar |
| 200 | c.200G>T | G67V | p.Gly67Val | Inactivating | FHH1 | 122257095 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 205 | c.205C>T | R69C | p.Arg69Cys | Inactivating | FHH1 | 122257100 | NA | SNV | Missense | Exon 3 | ECD | VUS | Greenberg | 2017 | NA | Greenberg H, Probst-Riordan J. Familial hypocalciuric hypercalcemia: A novel mutation. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 205 | c.205C>G | R69C | p.Arg69Cys | Inactivating | FHH1 (heterozygous) | 122257100 | NA | SNV | Missense | Exon 3 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 205 | c.205C>T | R69C | p.Arg69Cys | Inactivating | FHH1 | 122257100 | NA | SNV | Missense | Exon 3 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | FHH1 (heterozygous) | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Corrado | 2015 | 25828954 | 10.1002/jbmr.2516 | Journal article |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | NSHPT (compound heterozygous) | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Corrado | 2015 | 25828954 | 10.1002/jbmr.2516 | Journal article |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | FHH1 (heterozygous) | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Haider | 2022 | NA | 10.1159/000518849 | Conference abstract |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | FHH1 | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Khairi | 2020 | 32761341 | 10.1007/s12672-020-00394-2 | Journal article |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | FHH1 (heterozygous) | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Patrick-Esteve | 2015 | NA | 10.1097/JIM.0000000000000146 | Conference abstract |
| 206 | c.206G>A | R69H | p.Arg69His | Inactivating | NSHPT (homozygous) | 122257101 | rs193922432 | SNV | Missense | Exon 3 | ECD | Conflicting interpretations of pathogenicity | Wilhelm-Bals | 2012 | 22331334 | 10.1542/peds.2011-0128 | Journal article |
| 209 | c.209G>A | W70* | p.Trp70* | NA | NA | 122257104 | NA | SNV | Nonsense | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1177515/?oq=%22NM_000388.4(CASR):c.209G%3EA%20(p.Trp70Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.209G%3EA%20(p.Trp70Ter)#id_second | Clinvar |
| 212 | c.212T>G | L71* | p.Leu71* | Inactivating | FHH1 (heterozygous) | 122257107 | NA | SNV | Nonsense | Exon 3 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 212 | c.212T>G | L71* | p.Leu71* | Inactivating | FHH1 (heterozygous) | 122257107 | NA | SNV | Nonsense | Exon 3 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 220 | c.220A>G | M74V | p.Met74Val | Inactivating | FHH1 | 122257115 | NA | SNV | Missense | Exon 3 | ECD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 220 | c.220A>G | M74V | p.Met74Val | Inactivating | FHH1 | 122257115 | NA | SNV | Missense | Exon 3 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 222 | c.222_226delGATAT | M74Ifs*24 | p.Met74Ilefs*24 | Inactivating | FHH1 (heterozygous) | 122257117 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Atay | 2014 | 24735972 | 10.1016/j.bone.2014.04.010 | Journal article |
| 222 | c.222_226delGATAT | M74Ifs*24 | p.Met74Ilefs*24 | Inactivating | NSHPT (homozygous) | 122257117 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Atay | 2014 | 24735972 | 10.1016/j.bone.2014.04.010 | Journal article |
| 222 | c.222_226delGATAT | M74Ifs*24 | p.Met74Ilefs*24 | Inactivating | NSHPT (homozygous) | 122257117 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Çömlek | 2022 | 35399047 | 10.5114/pedm.2022.115070 | Journal article |
| 222 | c.222_226delGATAT | M74Ifs*24 | p.Met74Ilefs*24 | Inactivating | FHH1 (heterozygous) | 122257117 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Özgüç Çömlek | 2022 | 35399047 | 10.5114/pedm.2022.115070 | Journal article |
| 230 | c.230C>T | A77V | p.Ala77Val | Inactivating | FHH1 | 122257125 | NA | SNV | Missense | Exon 3 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 230 | c.230C>T | A77V | p.Ala77Val | Inactivating | FHH1 | 122257125 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 240 | c.240G>T | E80D | p.Glu80Asp | Inactivating | FHH1 | 122257135 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 242 | c.242T>A | I81K | p.Ile81Lys | Inactivating | FHH1 | 122257137 | NA | SNV | Missense | Exon 3 | ECD | NA | Joseph | 2022 | NA | https://jme.bioscientifica.com/abstract/journals/jme/69/1/JME-21-0263.xml | Journal article |
| 243 | c.243A>G | I81M | p.Ile81Met | Inactivating | FHH1 | 122257138 | NA | SNV | Missense | Exon 3 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 246 | c.246C>G | N82K | p.Asn82Lys | Inactivating | FHH1 | 122257141 | NA | SNV | Missense | Exon 3 | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 246 | c.246C>G | N82K | p.Asn82Lys | Inactivating | FHH1 | 122257141 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 272 | c.272T>A | L91* | p.Leu91* | Inactivating | FHH1 | 122257167 | NA | SNV | Nonsense | Exon 3 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 280 | c.280G>C | G94R | p.Gly94Arg | Inactivating | FHH1 | 122257175 | NA | SNV | Missense | Exon 3 | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 280 | c.280G>T | G94* | p.Gly94* | Inactivating | NSHPT (compound heterozygous) | 122257175 | rs104893705 | SNV | Nonsense | Exon 3 | ECD | VUS/Pathogenic | Ward | 2004 | 15292296 | 10.1210/jc.2003-031653 | Journal article |
| 281 | c.281G>A | G94E | p.Gly94Glu | Inactivating | NSHPT (homozygous) | 122257176 | rs1576854356 | SNV | Missense | Exon 3 | ECD | VUS | Solovitz | 2019 | NA | 10.1159/000501868 | Conference abstract |
| 284 | c.284A>G | Y95C | p.Tyr95Cys | Inactivating | FHH1 | 122257179 | rs1060502850 | SNV | Missense | Exon 3 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 295 | c.295G>C | D99H | p.Asp99His | Inactivating | NSHPT (homozygous) | 122257190 | NA | SNV | Missense | Exon 3 | ECD | NA | Haider | 2022 | NA | 10.1159/000518849 | Conference abstract |
| 295 | c.295G>C | D99H | p.Asp99His | Inactivating | NSHPT (homozygous) | 122257190 | NA | SNV | Missense | Exon 3 | ECD | NA | Shaukat | 2022 | NA | 10.47391/JPMA.4195 | Journal article |
| 299 | c.299C>T | T100I | p.Thr100Ile | Inactivating | FHH1 (heterozygous) | 122257194 | NA | SNV | Missense | Exon 3 | ECD | NA | Warner | 2004 | 14985373 | 10.1136/jmg.2003.016725 | Journal article |
| 301 | c.301T>C | C101R | p.Cys101Arg | Inactivating | FHH1 | 122257196 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 303 | c.303C>G | C101W | p.Cys101Trp | Inactivating | FHH1 (heterozygous) | 122257198 | NA | SNV | Missense | Exon 3 | ECD | NA | Cetani | 2019 | NA | Cetani F, Borsari S, Pardi E, Biagioni T, Bagattini B, Saponaro F, et al. Functional characterization of four mutations of the calcium sensing receptor gene identified in patients with familial hypocalciuric hypercalcemia type 1. Journal of Bone and Mineral Research. 2019;34:312. | Conference abstract |
| 303 | c.303C>G | C101W | p.Cys101Trp | Inactivating | FHH1 | 122257198 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 310 | c.310G>A | V104I | p.Val104Ile | Activating | ADH1 | 122257205 | rs2074564250 | SNV | Missense | Exon 3 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 319 | c.319G>A | A107T | p.Ala107Thr | Inactivating | FHH1 | 122257214 | NA | SNV | Missense | Exon 3 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 323 | c.323del | L108fs* | p.Leu108fs* | NA | NA | 122257217 | rs1559956616 | Small deletion | Frameshift | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/598322/?oq=%22NM_000388.4(CASR):c.323del%20(p.Leu108fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.323del%20(p.Leu108fs) | Clinvar |
| 328 | c.328G>A | A110T | p.Ala110Thr | Inactivating | FHH1 | 122257223 | NA | SNV | Missense | Exon 3 | ECD | NA | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 328 | c.328G>A | A110T | p.Ala110Thr | Inactivating | FHH1 | 122257223 | NA | SNV | Missense | Exon 3 | ECD | NA | Nakamura | 2013 | 23966241 | 10.1210/jc.2013-1974 | Journal article |
| 329 | c.329C>A | A110D | p.Ala110Asp | Inactivating | FHH1 (heterozygous) | 122257224 | rs1559956624 | SNV | Missense | Exon 3 | ECD | VUS | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 329 | c.329C>A | A110D | p.Ala110Asp | Inactivating | FHH1 (heterozygous) | 122257224 | NA | SNV | Missense | Exon 3 | ECD | VUS | Bletsis | 2022 | 36189134 | 10.1016/j.aace.2022.05.002 | Journal article |
| 344 | c.344_358delTTGCTCAAAACAAAA | A116_I120del | p.Ala116_I120del | Activating | ADH1 | 122257239 | NA | Small deletion | Deletion | Exon 3 | ECD | NA | Sozaeva | 2022 | NA | 10.24110/0031-403X-2022-101-6-186-191 | Journal article |
| 346 | c.346G>C | A116P | p.Ala116Pro | Activating | ADH1 | 122257241 | NA | SNV | Missense | Exon 3 | ECD | NA | Chapman | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 346 | c.346G>C | A116P | p.Ala116Pro | Activating | ADH1 | 122257241 | NA | SNV | Missense | Exon 3 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 346 | c.346G>A | A116T | p.Ala116Thr | Activating | ADH1 | 122257241 | rs104893691 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Baron | 1996 | 8733126 | 10.1093/hmg/5.5.601 | Journal article |
| 349 | c.349C>T | Q117* | p.Gln117* | NA | NA | 122257244 | NA | SNV | Nonsense | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1451604/?oq=%22NM_000388.4(CASR):c.349C%3ET%20(p.Gln117Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.349C%3ET%20(p.Gln117Ter)#id_second | Clinvar |
| 354 | c.354C>A | N118K | p.Asn118Lys | Activating | ADH1 | 122257249 | rs104893695 | SNV | Missense | Exon 3 | ECD | Pathogenic | Hu | 2002 | 12162500 | 10.1359/jbmr.2002.17.8.1461 | Journal article |
| 354 | c.354C>A | N118K | p.Asn118Lys | Activating | ADH1 | 122257249 | rs104893695 | SNV | Missense | Exon 3 | ECD | Pathogenic | Pearce | 1996 | 8813042 | 10.1056/NEJM199610103351505 | Journal article |
| 354 | c.354C>A | N118K | p.Asn118Lys | Activating | ADH1 | 122257249 | rs104893695 | SNV | Missense | Exon 3 | ECD | Pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 356 | c.356A>T | K119I | p.Lys119Ile | Activating | ADH1 | 122257251 | NA | SNV | Missense | Exon 3 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 357 | c.357_358del | K119fs* | p.Lys119fs* | NA | NA | 122257250 | NA | Small deletion | Frameshift | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1453750/?oq=%22NM_000388.4(CASR):c.357_358del%20(p.Lys119fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.357_358del%20(p.Lys119fs)#id_second | Clinvar |
| 361 | c.361_364delGATT | D121fs*2 | p.Asp121fs*2 | Inactivating | FHH1 (heterozygous) | 122257256 | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Falchetti | 2012 | 22315359 | 10.1530/EJE-11-0953 | Journal article |
| 365 | c.365C>G | S122C | p.Ser122Cys | Activating | ADH1 | 122257260 | NA | SNV | Missense | Exon 3 | ECD | NA | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 365 | c.365C>G | S122C | p.Ser122Cys | Activating | ADH1 | 122257260 | NA | SNV | Missense | Exon 3 | ECD | NA | Nakamura | 2013 | 23966241 | 10.1210/jc.2013-1974 | Journal article |
| 367 | c.367T>A | L123M | p.Leu123Met | Activating | ADH1 | 122257262 | NA | SNV | Missense | Exon 3 | ECD | VUS | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 368 | c.368T>C | L123S | p.Leu123Ser | Activating | ADH1 | 122257263 | NA | SNV | Missense | Exon 3 | ECD | NA | Festas Silva | 2021 | 34866060 | 10.1530/EDM-21-0005 | Journal article |
| 368 | c.368T>C | L123S | p.Leu123Ser | Activating | ADH1 | 122257263 | NA | SNV | Missense | Exon 3 | ECD | NA | Papadopoulou | 2017 | 28742508 | 10.14310/horm.2002.1734 | Journal article |
| 368 | c.368T>C | L123S | p.Leu123Ser | Activating | ADH1 | 122257263 | NA | SNV | Missense | Exon 3 | ECD | NA | Regala | 2015 | 27617113 | 10.1055/s-0035-1554979 | Journal article |
| 372 | c.372C>A | N124K | p.Asn124Lys | Activating | ADH1 | 122257267 | NA | SNV | Missense | Exon 3 | ECD | VUS | Ranieri | 2013 | 24244430 | 10.1371/journal.pone.0079113 | Journal article |
| 372 | c.372C>A | N124K | p.Asn124Lys | Activating | ADH1 | 122257267 | NA | SNV | Missense | Exon 3 | ECD | VUS | Schouten | 2011 | 21441391 | 10.1258/acb.2010.010139 | Journal article |
| 373 | c.373C>T | L125F | p.Leu125Phe | Activating | ADH1 | 122257268 | NA | SNV | Missense | Exon 3 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Hauche | 2000 | 11089548 | 10.1210/endo.141.11.7753 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Hu | 2002 | 12162500 | 10.1359/jbmr.2002.17.8.1461 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 with Bartter syndrome | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Letz | 2014 | 25506941 | 10.1371/journal.pone.0115178 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Sato | 2002 | 12107202 | 10.1210/jcem.87.7.8639 | Journal article |
| 374 | c.374T>C | L125P | p.Leu125Pro | Activating | ADH1 | 122257269 | NA | SNV | Missense | Exon 3 | ECD | Pathogenic | Vargas-Poussou | 2002 | 12191970 | 10.1097/01.asn.0000025781.16723.68 | Journal article |
| 377 | c.377A>T | D126V | p.Asp126Val | Activating | ADH1 | 122257272 | rs1553766257 | SNV | Missense | Exon 3 | ECD | VUS | Rasmussen | 2018 | 29743878 | 10.3389/fendo.2018.00200 | Journal article |
| 379 | c.379G>A | E127K | p.Glu127Lys | Activating | ADH1 | 122257274 | rs104893710 | SNV | Missense | Exon 3 | ECD | Pathogenic | Hawkes | 2020 | 33112267 | 10.1530/EJE-20-0710 | Journal article |
| 379 | c.379G>A | E127K | p.Glu127Lys | Activating | ADH1 | 122257274 | rs104893710 | SNV | Missense | Exon 3 | ECD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 379 | c.379G>A | E127K | p.Glu127Lys | Activating | ADH1 | 122257274 | rs104893710 | SNV | Missense | Exon 3 | ECD | Pathogenic | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 380 | c.380A>G | E127G | p.Glu127Gly | Activating | ADH1 | 122257275 | rs121909260 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 380 | c.380A>C | E127A | p.Glu127Ala | Activating | ADH1 | 122257275 | rs121909260 | SNV | Missense | Exon 3 | ECD | Pathogenic | Loseva | 2018 | NA | Loseva V, Mulloy AL, Sinnott BP. Autosomal dominant hypocalcemia: An important but often unrecognized cause of hypoparathyroidism. Endocrine Reviews. 2018;39(2). | Conference abstract |
| 380 | c.380A>C | E127A | p.Glu127Ala | Activating | ADH1 | 122257275 | rs121909260 | SNV | Missense | Exon 3 | ECD | Pathogenic | Pollak | 1994 | 7874174 | 10.1038/ng1194-303 | Journal article |
| 382 | c.382T>C | F128L | p.Phe128Leu | Activating | ADH1 | 122257277 | rs104893696 | SNV | Missense | Exon 3 | ECD | Pathogenic | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 383 | c.383T>G | F128C | p.Phe128Cys | Activating | ADH1 | 122257278 | NA | SNV | Missense | Exon 3 | ECD | NA | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 384 | c.384C>A | F128L | p.Phe128Leu | Activating | ADH1 | 122257279 | rs1553766262 | SNV | Missense | Exon 3 | ECD | Pathogenic | Pearce | 1996 | 8813042 | 10.1056/NEJM199610103351505 | Journal article |
| 385 | c.385T>A | C129S | p.Cys129Ser | Activating | ADH1 | 122257280 | NA | SNV | Missense | Exon 3 | ECD | NA | Hirai | 2001 | 11289719 | 10.1007/s100380170124 | Journal article |
| 385 | c.385T>C | C129R | p.Cys129Arg | Activating | ADH1 | 122257280 | NA | SNV | Missense | Exon 3 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 386 | c.386G>A | C129Y | p.Cys129Tyr | Activating | ADH1 | 122257281 | rs2074565392 | SNV | Missense | Exon 3 | ECD | Pathogenic | Burren | 2005 | 16128246 | 10.1515/jpem.2005.18.7.689 | Journal article |
| 386 | c.386G>C | C129S | p.Cys129Ser | Activating | ADH1 | 122257281 | NA | SNV | Missense | Exon 3 | ECD | NA | Kurozumi | 2013 | 24042516 | 10.2169/internalmedicine.52.8375 | Journal article |
| 386 | c.386G>T | C129F | p.Cys129Phe | Activating | ADH1 | 122257281 | NA | SNV | Missense | Exon 3 | ECD | NA | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 386 | c.386G>A | C129Y | p.Cys129Tyr | Activating | ADH1 | 122257281 | rs2074565392 | SNV | Missense | Exon 3 | ECD | Pathogenic | Schouten | 2011 | 21441391 | 10.1258/acb.2010.010139 | Journal article |
| 391 | c.391T>A | C131R | p.Cys131Arg | Activating | ADH1 | 122257286 | NA | SNV | Missense | Exon 3 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 392 | c.392G>A | C131Y | p.Cys131Tyr | Activating | ADH1 | 122257287 | NA | SNV | Missense | Exon 3 | ECD | NA | Chen | 2017 | NA | 10.1159/000481424 | Conference abstract |
| 392 | c.392G>A | C131Y | p.Cys131Tyr | Activating | ADH1 | 122257287 | NA | SNV | Missense | Exon 3 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 392 | c.392G>T | C131F | p.Cys131Phe | Activating | ADH1 | 122257287 | NA | SNV | Missense | Exon 3 | ECD | NA | Suzuki | 2005 | 15960151 | 10.5414/CNP63481 | Journal article |
| 392 | c.392G>A | C131Y | p.Cys131Tyr | Activating | ADH1 | 122257287 | NA | SNV | Missense | Exon 3 | ECD | NA | Thim | 2014 | 25039540 | 10.1111/apa.12743 | Conference abstract |
| 392 | c.393C>G | C131W | p.Cys131Trp | Activating | ADH1 with Bartter syndrome | 122257288 | rs121909267 | SNV | Missense | Exon 3 | ECD | Pathogenic | Watanabe | 2002 | 12241879 | 10.1016/S0140-6736(02)09842-2 | Journal article |
| 393 | c.393C>G | C131W | p.Cys131Trp | Activating | ADH1 with Bartter syndrome | 122257288 | rs121909267 | SNV | Missense | Exon 3 | ECD | Pathogenic | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 393 | c.393C>G | C131W | p.Cys131Trp | Activating | ADH1 with Bartter syndrome | 122257288 | rs121909267 | SNV | Missense | Exon 3 | ECD | Pathogenic | Letz | 2014 | 25506941 | 10.1371/journal.pone.0115178 | Journal article |
| 398 | c.398A>T | E133V | p.Glu133Val | Activating | ADH1 | 122257293 | NA | SNV | Missense | Exon 3 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 398 | c.398A>T | E133V | p.Glu133Val | Activating | ADH1 | 122257293 | NA | SNV | Missense | Exon 3 | ECD | NA | Tan | 2022 | 35870823 | 10.1016/j.kint.2022.02.014 | Conference abstract |
| 407 | c.407C>T | P136L | p.Pro136Leu | Activating | ADH1 | 122257302 | NA | SNV | Missense | Exon 3 | ECD | NA | Finn | 2012 | NA | 10.1136/archdischild-2012-301885.291 | Journal article |
| 407 | c.407C>T | P136L | p.Pro136Leu | Activating | ADH1 | 122257302 | NA | SNV | Missense | Exon 3 | ECD | NA | Gupta | 2012 | NA | 10.1159/000343182 | Conference abstract |
| 408 | c.408C>T | P136L | p.Pro136Leu | Activating | ADH1 | 122257303 | NA | SNV | Missense | Exon 3 | ECD | NA | Baran | 2015 | 25766501 | 10.1016/j.mce.2015.02.021 | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 (heterozygous) | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Alam | 2021 | 35300448 | 10.4103/ijem.ijem_349_21 | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 (heterozygous) | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 (heterozygous) | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 (heterozygous) | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Rosen | 2017 | NA | Rosen SG. Pregnancy complicated by hypocalciuric hypercalcemia. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 (heterozygous) | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Chou | 1995 | 7726161 | NA | Journal article |
| 413 | c.413C>T | T138M | p.Thr138Met | Inactivating | FHH1 | 122257308 | rs121909263 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 416 | c.416T>C | I139T | p.Ile139Thr | Activating | ADH1 | 122257311 | rs1060502860 | SNV | Missense | Exon 3 | ECD | VUS | Guo | 2022 | 36935580 | 10.1515/jpem-2022-0623 | Journal article |
| 416 | c.416T>C | I139T | p.Ile139Thr | Activating | ADH1 | 122257311 | rs1060502860 | SNV | Missense | Exon 3 | ECD | VUS | Wu | 2022 | 35818129 | 10.1177/03000605221110489 | Journal article |
| 416 | c.416T>C | I139T | p.Ile139Thr | Activating | ADH1 | 122257311 | rs1060502860 | SNV | Missense | Exon 3 | ECD | VUS | Zung | 2023 | 36812896 | 10.1159/000529833 | Journal article |
| 419 | c.419C>T | A140V | p.Ala140Val | Inactivating | FHH1 | 122257314 | NA | SNV | Missense | Exon 3 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 419 | c.419C>T | A140V | p.Ala140Val | Inactivating | FHH1 | 122257314 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 422 | c.422T>C | V141A | p.Val141Ala | Inactivating | FHH1 | 122257317 | rs1085307643 | SNV | Missense | Exon 3 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 427 | c.427G>A | G143R | p.Gly143Arg | Inactivating | FHH1 (heterozygous) | 122257322 | rs769256610 | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 427 | c.427G>C | G143R | p.Gly143Arg | Inactivating | FHH1 | 122257322 | NA | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 427 | c.427G>C | G143R | p.Gly143Arg | Inactivating | FHH1 | 122257322 | NA | SNV | Missense | Exon 3 | ECD | Likely pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 428 | c.428G>A | G143E | p.Gly143Glu | Inactivating | FHH1 (heterozygous) | 122257323 | rs121909264 | SNV | Missense | Exon 3 | ECD | Pathogenic/Likely pathogenic | Chou | 1995 | 7726161 | NA | Journal article |
| 434 | c.434C>T | T145I | p.Thr145Ile | Inactivating | FHH1 | 122257329 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 437 | c.437G>A | G146D | p.Gly146Asp | Inactivating | FHH1 (heterozygous) | 122257332 | NA | SNV | Missense | Exon 3 | ECD | VUS | Carllson | 2019 | 30895164 | 10.1155/2019/9468252 | Journal article |
| 439 | c.439C>T | S147L | p.Ser147Leu | Inactivating | FHH1 (heterozygous) | 122257334 | NA | SNV | Missense | Exon 3 | ECD | NA | Majumdar | 2018 | 33062349 | 10.1155/2020/8752610 | Journal article |
| 448 | c.448_449dup | T151fs* | p.Thr151fs* | NA | NA | 122257340 | rs1576854561 | Small insertion | Frameshift | Exon 3 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/664654/?oq=%22NM_000388.4(CASR):c.448_449dup%20(p.Thr151fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.448_449dup%20(p.Thr151fs)#id_second | Clinvar |
| 449 | c.449C>G | S150C | p.Ser150Cys | Inactivating | FHH1 | 122257344 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 452 | c.452C>T | T151M | p.Thr151Met | Activating | ADH1 | 122257347 | rs104893694 | SNV | Missense | Exon 3 | ECD | Pathogenic | Gonzales | 2013 | 23186954 | 10.4158/EP12132.CR | Journal article |
| 452 | c.452C>T | T151M | p.Thr151Met | Activating | ADH1 | 122257347 | rs104893694 | SNV | Missense | Exon 3 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 452 | c.452C>G | T151R | p.Thr151Arg | Activating | ADH1 | 122257347 | NA | SNV | Missense | Exon 3 | ECD | NA | Letz | 2010 | 20668040 | 10.1210/jc.2010-0651 | Journal article |
| 452 | c.452C>T | T151M | p.Thr151Met | Activating | ADH1 | 122257347 | rs104893694 | SNV | Missense | Exon 3 | ECD | Pathogenic | Pearce | 1996 | 8813042 | 10.1056/NEJM199610103351505 | Journal article |
| 452 | c.452C>T | T151M | p.Thr151Met | Activating | ADH1 | 122257347 | rs104893694 | SNV | Missense | Exon 3 | ECD | Pathogenic | Sørheim | 2010 | 20501971 | 10.1159/000303188 | Journal article |
| 452 | c.452C>G | T151R | p.Thr151Arg | Activating | ADH1 | 122257347 | NA | SNV | Missense | Exon 3 | ECD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 472 | c.472G>A | G158R | p.Gly158Arg | Inactivating | FHH1 | 122257367 | NA | SNV | Missense | Exon 3 | ECD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 473 | c.473G>A | G158E | p.Gly158Glu | Inactivating | FHH1 (heterozygous) | 122257368 | rs2074566747 | SNV | Missense | Exon 3 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 473 | c.473G>C | G158A | p.Gly158Ala | Inactivating | FHH1 (heterozygous) | 122257368 | NA | SNV | Missense | Exon 3 | ECD | NA | Porta Vilaró | 2022 | NA | 10.1016/j.mcpsp.2021.100260 | Journal article |
| 476 | c.476T>G | L159R | p.Leu159Arg | Inactivating | FHH1 (heterozygous) | 122257371 | NA | SNV | Missense | Exon 3 | ECD | NA | Ho | 2010 | 21175100 | 10.1515/jpem.2010.156 | Journal article |
| 476 | c.476T>C | L159P | p.Leu159Pro | Inactivating | FHH1 (heterozygous) | 122257371 | NA | SNV | Missense | Exon 3 | ECD | NA | Simonds | 2002 | 11807402 | 10.1097/00005792-200201000-00001 | Journal article |
| 482 | c.482A>G | Y161C | p.Tyr161Cys | Inactivating | FHH1 (heterozygous) | 122257377 | NA | SNV | Missense | Exon 3 | ECD | NA | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 482 | c.482A>G | Y161C | p.Tyr161Cys | Inactivating | FHH1 (heterozygous) | 122257377 | NA | SNV | Missense | Exon 3 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 484 | c.484A>T | I162F | p.Ile162Phe | Inactivating | FHH1 | 122257379 | NA | SNV | Missense | Exon 3 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 488 | c.488C>T | P163L | p.Pro163Leu | Inactivating | FHH1 (heterozygous) | 122257383 | NA | SNV | Missense | Exon 3 | ECD | NA | Cetani | 2019 | NA | Cetani F, Borsari S, Pardi E, Biagioni T, Bagattini B, Saponaro F, et al. Functional characterization of four mutations of the calcium sensing receptor gene identified in patients with familial hypocalciuric hypercalcemia type 1. Journal of Bone and Mineral Research. 2019;34:312. | Conference abstract |
| 488 | c.488C>G | P163R | p.Pro163Arg | Inactivating | FHH1 (heterozygous) | 122257383 | NA | SNV | Missense | Exon 3 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 490 | c.490C>T | Q164* | p.Gln164* | Inactivating | NSHPT (homozygous) | 122257385 | NA | SNV | Nonsense | Exon 3 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 490 | c.490C>A | Q164R | p.Gln164Arg | Inactivating | FHH1 (heterozygous) | 122257385 | rs2074567065 | SNV | Missense | Exon 3 | ECD | VUS | Kim | 2014 | 24754691 | 10.1111/imj.12383 | Journal article |
| 490 | c.490C>T | Q164* | p.Gln164* | Inactivating | FHH1 | 122257385 | NA | SNV | Nonsense | Exon 3 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 490 | c.490C>T | Q164* | p.Gln164* | Inactivating | NSHPT (homozygous) | 122257385 | NA | SNV | Nonsense | Exon 3 | ECD | VUS | Waller | 2004 | 15241688 | 10.1007/s00431-004-1491-0 | Journal article |
| 491 | c.491A>G | Q164R | p.Gln164Arg | Inactivating | FHH1 (heterozygous) | 122257386 | rs2074567065 | SNV | Missense | Exon 3 | ECD | VUS | Coughlan | 2022 | 35420006 | Coughlan AK, Khan F, Brassill MJ. A Novel Genetic Variant Resulting in Familial Hypocalciuric Hypercalcaemia. Irish Medical Journal. 2022;115(2). | Journal article |
| 492 | c.492+1G>C | NA | NA | Inactivating | NSHPT (homozygous) | 122257388 | NA | SNV | Splice donor | Exon 3-intron 3 splice site | ECD | NA | Sadacharan | 2020 | 32699790 | 10.4103/ijem.IJEM_53_20 | Journal article |
| 493 | c.493-12G>A | NA | NA | Inactivating | NSHPT (homozygous) | 122261516 | NA | SNV | Splice acceptor | Intron 3-Exon 4 splice site | ECD | NA | Arif | 2021 | NA | https://ecommons.aku.edu/pakistan_fhs_mc_women_childhealth_paediatr/1306/ | Conference abstract |
| 493 | c.493-1G>A | Two amino acid missense peptide followed by a stop at codon 167 or an in-frame deletion of exon 4 | Two amino acid missense peptide followed by a stop at codon 167 or an in-frame deletion of exon 4 | Inactivating | FHH1 (heterozygous) | 122261527 | NA | SNV | Splice acceptor | Intron 3-Exon 4 splice site | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 493 | c.493_497del | V165fs*24 | p.Val165fs*24 | Inactivating | NSHPT (homozygous) | 122261528 | NA | Small deletion | Frameshift | Exon 4 | ECD | NA | Ward | 2006 | 16509534 | 10.1515/jpem.2006.19.1.93 | Journal article |
| 496 | c.496A>G | S166G | p.Ser166Gly | Inactivating | FHH1 | 122261531 | rs193922441 | SNV | Missense | Exon 4 | ECD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 497 | c.497G>T | S166I | p.Ser166Ile | Inactivating | FHH1 | 122261532 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 501 | c.501T>A | Y167* | p.Tyr167* | Inactivating | FHH1 | 122261536 | NA | SNV | Nonsense | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1697234/?oq=%22NM_000388.4(CASR):c.501T%3EA%20(p.Tyr167Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.501T%3EA%20(p.Tyr167Ter)#id_second | Clinvar |
| 503 | c.503C>T | A168V | p.Ala168Val | Inactivating | FHH1 | 122261538 | rs1576857840 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/801995/?oq=%22NM_000388.4(CASR):c.503C%3ET%20(p.Ala168Val)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.503C%3ET%20(p.Ala168Val) | Clinvar |
| 505 | c.505T>C | S169P | p.Ser169Pro | Inactivating | FHH1 | 122261540 | rs1553766709 | SNV | Missense | Exon 4 | ECD | VUS | Khairi | 2020 | 32761341 | 10.1007/s12672-020-00394-2 | Journal article |
| 512 | c.512G>A | S171N | p.Ser171Asn | Inactivating | FHH1 (heterozygous) | 122261547 | NA | SNV | Missense | Exon 4 | ECD | NA | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 512 | c.512G>A | S171N | p.Ser171Asn | Inactivating | FHH1 (heterozygous) | 122261547 | NA | SNV | Missense | Exon 4 | ECD | NA | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 512 | c.512G>A | S171N | p.Ser171Asn | Inactivating | FHH1 (heterozygous) | 122261547 | NA | SNV | Missense | Exon 4 | ECD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 512 | c.512G>A | S171N | p.Ser171Asn | Inactivating | FHH1 (heterozygous) | 122261547 | NA | SNV | Missense | Exon 4 | ECD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 513 | c.513C>A | S171R | p.Ser171Arg | Inactivating | FHH1 | 122261548 | rs764149433 | SNV | Missense | Exon 4 | ECD | VUS | Khairi | 2020 | 32761341 | 10.1007/s12672-020-00394-2 | Journal article |
| 513 | c.513C>A | S171R | p.Ser171Arg | Inactivating | FHH1 | 122261548 | rs764149433 | SNV | Missense | Exon 4 | ECD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 513 | c.513C>A | S171R | p.Ser171Arg | Inactivating | FHH1 | 122261548 | rs764149433 | SNV | Missense | Exon 4 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 514 | c.514A>G | R172G | p.Arg172Gly | Inactivating | FHH1 (heterozygous) | 122261549 | rs201851934 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 514 | c.514A>G | R172G | p.Arg172Gly | Inactivating | FHH1 (heterozygous) | 122261549 | rs201851934 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hinnie | 2009 | 20034274 | 10.1258/rsmsmj.54.4.11 | Journal article |
| 514 | c.514A>G | R172G | p.Arg172Gly | Inactivating | FHH1 | 122261549 | rs201851934 | SNV | Missense | Exon 4 | ECD | Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 514 | c.514A>G | R172G | p.Arg172Gly | Inactivating | FHH1 | 122261549 | rs201851934 | SNV | Missense | Exon 4 | ECD | Pathogenic | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 514 | c.514A>G | R172G | p.Arg172Gly | Inactivating | FHH1 | 122261549 | rs201851934 | SNV | Missense | Exon 4 | ECD | Pathogenic | Nakamura | 2013 | 23966241 | 10.1210/jc.2013-1974 | Journal article |
| 517 | c.517C>T | L173F | p.Leu173Phe | Activating | ADH1 | 122261552 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 518 | c.518T>C | L173P | p.Leu173Pro | Inactivating | FHH1 (heterozygous) | 122261553 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic | Felderbauer | 2005 | 15662592 | 10.1055/s-2004-830523 | Journal article |
| 518 | c.518T>C | L173P | p.Leu173Pro | Inactivating | FHH1 | 122261553 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 521 | c.521T>G | L174R | p.Leu174Arg | Inactivating | FHH1 (heterozygous) | 122261556 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 521 | c.521T>G | L174R | p.Leu174Arg | Inactivating | FHH1 (heterozygous) | 122261556 | NA | SNV | Missense | Exon 4 | ECD | NA | Ward | 1997 | 9298824 | 10.1002/(SICI)1098-1004(1997)10:3<233::AID-HUMU9>3.0.CO;2-J | Journal article |
| 521 | c.521T>G | L174R | p.Leu174Arg | Inactivating | FHH1 (heterozygous) | 122261556 | NA | SNV | Missense | Exon 4 | ECD | NA | Ward | 2006 | 16649980 | 10.1111/j.1365-2265.2006.02512.x | Journal article |
| 524 | c.524G>T | S175I | p.Ser175Ile | Inactivating | FHH1 | 122261559 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 528 | c.528del | N176fs* | p.Asn176fs* | NA | NA | 122261563 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1378885/?oq=%22NM_000388.4(CASR):c.528del%20(p.Asn176fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.528del%20(p.Asn176fs)#id_second | Clinvar |
| 532 | c.532A>G | N178D | p.Asn178Asp | Inactivating | FHH1 (heterozygous) | 122261567 | rs1060502855 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Pearce | 1996 | 9039332 | 10.1046/j.1365-2265.1996.750891.x | Journal article |
| 532 | c.532A>G | N178D | p.Asn178Asp | Inactivating | FHH1 (heterozygous) | 122261567 | rs1060502855 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Saeed | 2018 | NA | Saeed ZI, Lteif AA. A variance of the calcium sensing receptor (CASR) gene becoming clinically significant. Endocrine Reviews. 2018;39(2). | Journal article |
| 532 | c.532A>T | N178Y | p.Asn178Tyr | Activating | ADH1 | 122261567 | NA | SNV | Missense | Exon 4 | ECD | VUS | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 539 | c.539T>G | F180C | p.Phe180Cys | Inactivating | FHH1 | 122261574 | rs121909268 | SNV | Missense | Exon 4 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 539 | c.539T>G | F180C | p.Phe180Cys | Inactivating | FHH1 | 122261574 | rs121909268 | SNV | Missense | Exon 4 | ECD | Pathogenic | Zajíčková | 2007 | 17473068 | 10.1210/jc.2007-0123 | Journal article |
| 547 | c.547T>C | F183L | p.Phe183Leu | Inactivating | FHH1 | 122261582 | NA | SNV | Missense | Exon 4 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 547 | c.547_548del | F183fs* | p.Phe183fs* | NA | NA | 122261581 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1075778/?oq=%22NM_000388.4(CASR):c.547_548del%20(p.Phe183fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.547_548del%20(p.Phe183fs)#id_second | Clinvar |
| 553 | c.553C>T | R185* | p.Arg185* | Inactivating | NSHPT (compound heterozygous) | 122261588 | rs104893707 | SNV | Nonsense | Exon 4 | ECD | VUS/Likely pathogenic/Pathogenic | Kobayashi | 1997 | 9253359 | 10.1210/jcem.82.8.4135 | Journal article |
| 553 | c.553C>G | R185G | p.Arg185Gly | Inactivating | FHH1 | 122261588 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Aubert-Mucca | 2015 | 34169121 | 10.1016/j.bonr.2021.101097 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Bond | 2017 | NA | Bond DT, Paulus AO. Severe hypercalcemia in a patient with familial hypocalciuric hypercalcemia with a rare calcium-sensing receptor (CASR) R185Q mutation treated with cinacalcet. Endocrine Reviews. 2017;38(3) | Conference abstract |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Chapman | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Döğer | 2019 | NA | https://abstracts.eurospe.org/hrp/0092/hrp0092p2-44 | Conference abstract |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Fisher | 2015 | 26161261 | 10.1530/EDM-15-0040 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Forman | 2018 | 30730839 | 10.1515/jpem-2018-0307 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Gannon | 2013 | 24203066 | 10.1210/jc.2013-2834 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Glaudo | 2016 | 27666534 | 10.1530/EJE-16-0223 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Höppner | 2021 | 34659108 | 10.3389/fendo.2021.700612 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Krupinova | 2020 | 33369373 | 10.14341/probl12537 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Moll | 2013 | NA | Moll GW, Moll CL. Progressive hypercalcemia in a prepubertal male: Assessment and response to cinacalcet. Endocrine Reviews. 2013;34(3). | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Obermannova | 2009 | 18751724 | 10.1007/s00431-008-0794-y | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Palmieri | 2022 | 35141253 | 10.3389/fmed.2021.809067 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Pollak | 1993 | 7916660 | 10.1016/0092-8674(93)90617-y | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | NSHPT (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Reh | 2011 | 21289269 | 10.1210/jc.2010-1306 | Journal article |
| 554 | c.554G>A | R185Q | p.Arg185Gln | Inactivating | FHH1 (heterozygous) | 122261589 | rs104893689 | SNV | Missense | Exon 4 | ECD | Pathogenic | Sánchez | 2019 | NA | Sánchez A, Rossi E. Genetic hypercalcemic hypocalciuria in a Prepubertal boy. Revista Medica de Rosario. 2019;85(2):77-80. | Journal article |
| 554 | c.554del | R185fs* | p.Arg185fs* | Inactivating | FHH1 | 122261589 | rs193922442 | Small deletion | Frameshift | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35801/?oq=%22NM_000388.4(CASR):c.554del%20(p.Arg185fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.554del%20(p.Arg185fs)#id_second | Clinvar |
| 557 | c.557C>A | T186N | p.Thr186Asn | Activating | ADH1 | 122261592 | NA | SNV | Missense | Exon 4 | ECD | NA | Tsuji | 2021 | 33506158 | 10.1210/jendso/bvaa190 | Journal article |
| 568 | c.568G>A | D190N | p.Asp190Asn | Inactivating | FHH1 | 122261603 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 569 | c.569A>G | D190G | p.Asp190Gly | Inactivating | FHH1 (heterozygous) | 122261604 | NA | SNV | Missense | Exon 4 | ECD | NA | Nanjo | 2010 | 20697181 | 10.1507/endocrj.k10e-178 | Journal article |
| 570 | c.570delT | D190fs*67 | p.Asp190fs*67 | Inactivating | NSHPT (homozygous) | 122261605 | NA | Small deletion | Frameshift | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 570 | c.570T>G | D190E | p.Asp190Glu | Inactivating | FHH1 | 122261605 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 570 | c.570delT | D190fs*67 | p.Asp190fs*67 | Inactivating | NSHPT (homozygous) | 122261605 | NA | Small deletion | Frameshift | Exon 4 | ECD | NA | Webb | 2009 | NA | 10.1016/j.bone.2009.04.137 | Journal article |
| 571 | c.571G>A | E191K | p.Glu191Lys | Activating | ADH1 | 122261606 | rs104893697 | SNV | Missense | Exon 4 | ECD | Pathogenic | Pearce | 1996 | 8813042 | 10.1056/NEJM199610103351505 | Journal article |
| 577 | c.577C>T | Q193* | p.Gln193* | NA | NA | 122261612 | rs1064793992 | SNV | Nonsense | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/419616/?oq=%22NM_000388.4(CASR):c.577C%3ET%20(p.Gln193Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.577C%3ET%20(p.Gln193Ter)#id_second | Clinvar |
| 593 | c.593C>A | A198E | p.Ala198Glu | Inactivating | FHH1 | 122261628 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 613 | c.613C>T | R205C | p.Arg205Cys | Activating | ADH1 | 122261648 | rs775751453 | SNV | Missense | Exon 4 | ECD | VUS | Ji | 2021 | 34160437 | 10.1097/MD.0000000000026443 | Journal article |
| 623 | c.623G>C | W208S | p.Trp208Ser | Inactivating | FHH1 (heterozygous) | 122261658 | rs2074623917 | SNV | Missense | Exon 4 | ECD | VUS | Demedts | 2008 | 18296474 | 10.1164/ajrccm.177.5.558 | Journal article |
| 623 | c.623G>C | W208S | p.Trp208Ser | Inactivating | FHH1 (heterozygous) | 122261658 | rs2074623917 | SNV | Missense | Exon 4 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 624 | c.624G>C | W208G | p.Trp208Gly | Inactivating | FHH1 (heterozygous) | 122261659 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 635 | c.635T>G | I212S | p.Ile212Ser | Inactivating | NSHPT (homozygous) | 122261670 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 635 | c.635T>C | I212T | p.Ile212Thr | Inactivating | FHH1 (heterozygous) | 122261670 | rs1576858127 | SNV | Missense | Exon 4 | ECD | VUS | Marcocci | 2003 | 14602739 | 10.1210/jc.2003-030739 | Journal article |
| 638 | c.638C>A | A213E | p.Ala213Glu | Inactivating | FHH1 (heterozygous) | 122261673 | NA | SNV | Missense | Exon 4 | ECD | NA | Elamin | 2010 | 21034470 | 10.1186/1752-1947-4-349 | Journal article |
| 643 | c.643G>C | D215H | p.Asp215His | Inactivating | FHH1 | 122261678 | rs1553731681 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35802/?oq=%22NM_000388.4(CASR):c.643G%3EC%20(p.Asp215His)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.643G%3EC%20(p.Asp215His)#id_second | Clinvar |
| 644 | c.644A>G | D215G | p.Asp215Gly | Inactivating | FHH1 (heterozygous) | 122261679 | NA | SNV | Missense | Exon 4 | ECD | NA | Marstrand | 2021 | 34556169 | 10.1186/s13256-021-03051-6 | Journal article |
| 649 | c.649G>T | D217Y | p.Asp217Tyr | Inactivating | FHH1 | 122261684 | rs201091657 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/430465/?oq=%22NM_000388.4(CASR):c.649G%3ET%20(p.Asp217Tyr)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.649G%3ET%20(p.Asp217Tyr)#id_first | Clinvar |
| 652 | c.652T>C | Y218H | p.Tyr218His | Inactivating | FHH1 (heterozygous) | 122261687 | rs1057520583 | SNV | Missense | Exon 4 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 652 | c.652T>A | Y218N | p.Tyr218Asn | Inactivating | FHH1 | 122261687 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 652 | c.652T>G | Y218D | p.Tyr218Asp | NA | NA | 122261687 | rs1057520583 | SNV | Missense | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/379352/?oq=%22NM_000388.4(CASR):c.652T%3EG%20(p.Tyr218Asp)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.652T%3EG%20(p.Tyr218Asp)#id_second | Clinvar |
| 653 | c.653A>G | Y218C | p.Tyr218Cys | Inactivating | FHH1 (heterozygous) | 122261688 | rs2074624616 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Cetani | 2003 | 12580936 | 10.1046/j.1365-2265.2003.01696.x | Journal article |
| 653 | c.653A>G | Y218C | p.Tyr218Cys | Inactivating | FHH1 | 122261688 | rs2074624616 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 653 | c.653A>G | Y218C | p.Tyr218Cys | Inactivating | FHH1 | 122261688 | rs2074624616 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 653 | c.653A>G | Y218C | p.Tyr218Cys | Inactivating | FHH1 (heterozygous) | 122261688 | rs2074624616 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Quaglia | 2012 | NA | 10.1093/ndt/gfs201 | Conference abstract |
| 653 | c.653A>C | Y218S | p.Tyr218Ser | Inactivating | FHH1 (heterozygous) | 122261688 | NA | SNV | Missense | Exon 4 | ECD | NA | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | NSHPT (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Fox | 2014 | 17974727 | 10.1542/peds.2006-3209 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | NSHPT (homozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | NSHPT (homozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Orlova | 2011 | NA | 10.1159/000334328 | Conference abstract |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Park | 2022 | 35586626 | 10.3389/fendo.2022.853171 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Rasmussen | 2011 | 22142470 | 10.1186/1752-1947-5-564 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Schwarz | 2000 | 10885494 | 10.1080/003655100750044875 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 (heterozygous) | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Simonds | 2002 | 11807402 | 10.1097/00005792-200201000-00001 | Journal article |
| 658 | c.658C>T | R220W | p.Arg220Trp | Inactivating | FHH1 | 122261693 | rs1482119762 | SNV | Missense | Exon 4 | ECD | Pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Alon | 2010 | 20495831 | 10.1007/s00467-010-1547-5 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Bahíllo-Curieses | 2020 | 32115198 | 10.1016/j.medcli.2020.01.005 | Journal article |
| 659 | c.659G>C | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | NA | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Egan | 2012 | 23081733 | 10.1007/s00774-012-0399-4 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Fukumoto | 2001 | 11763315 | 10.1097/00019606-200112000-00006 | Journal article |
| 659 | c.659G>C | R220P | p.Arg220Pro | Inactivating | FHH1 (heterozygous) | 122261694 | NA | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Mohammed | 2008 | 18830196 | https://medscimonit.com/abstract/index/idArt/869397 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Pearce | 1996 | 9039332 | 10.1046/j.1365-2265.1996.750891.x | Journal article |
| 659 | c.659G>A | R220Q | p.Arg220Gln | Inactivating | FHH1 (heterozygous) | 122261694 | rs1202110240 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Ziv | 2019 | NA | 10.1159/000501868 | Conference abstract |
| 661 | c.661C>T | P221S | p.Pro221Ser | Inactivating | FHH1 | 122261696 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 661 | c.661C>T | P221S | p.Pro221Ser | Inactivating | FHH1 (heterozygous) | 122261696 | NA | SNV | Missense | Exon 4 | ECD | NA | Pearce | 1996 | 9039332 | 10.1046/j.1365-2265.1996.750891.x | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Bastida | 2021 | NA | Bastida MG, Servidio M, Marino R, Avila S, Viterbo G, Boquete H. Autosomal dominant hypocalcemia: Description of clinical and biochemical findings in a family with the missense pathogenic variant p. pro221leu in the casr gene. Revista Argentina de Endocrinologia y Metabolismo. 2021;58(SUPPL 1):46. | Conference abstract |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Chikatsu | 2003 | 12733714 | 10.1507/endocrj.50.91 | Journal article |
| 662 | c.662C>A | P221Q | p.Pro221Gln | Inactivating | FHH1 (heterozygous) | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 662 | c.662C>A | P221Q | p.Pro221Gln | Inactivating | FHH1 (heterozygous) | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Conley | 2000 | 11136551 | 10.1006/mgme.2000.3096 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic | Eom | 2011 | NA | Eom YS, Choi B, Yi HS, Chung YS, Jung TS, Park SY, et al. Mutational analyses of genes in korean patients with familial or sporadic forms of isolated hypoparathyroidism: A series of korean hypopara registry study. Endocrine Reviews. 2011;32(3). | Conference abstract |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Guarnieri | 2012 | 22789683 | 10.1016/j.ymgme.2012.06.012 | Journal article |
| 662 | c.662C>A | P221Q | p.Pro221Gln | Inactivating | FHH1 (heterozygous) | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 662 | c.662C>A | P221Q | p.Pro221Gln | Inactivating | FHH1 (heterozygous) | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Khaliel | 2022 | NA | 10.1111/dmcn.15123 | Conference abstract |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Kim | 2010 | 20119591 | 10.3346/jkms.2010.25.2.317 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Letz | 2010 | 20668040 | 10.1210/jc.2010-0651 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 662 | c.662C>A | P221Q | p.Pro221Gln | Inactivating | FHH1 (heterozygous) | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Shibata | 2013 | NA | Shibata H, Hori N, Yoshida M, Mitui T, Narumi S, Hasegawa T. A family with autosomal dominant hypocalcaemia with sensorineural hearing impairment and low urine excretion of calcium. Hormone Research in Paediatrics. 2013;80:87. | Conference abstract |
| 662 | c.662C>T | P221L | p.Pro221Leu | Activating | ADH1 | 122261697 | rs397514728 | SNV | Missense | Exon 4 | ECD | Pathogenic | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 664 | c.664G>A | G222R | p.Gly222Arg | NA | NA | 122261699 | NA | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1331538/?oq=%22NM_000388.4(CASR):c.664G%3EA%20(p.Gly222Arg)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.664G%3EA%20(p.Gly222Arg)#id_second | Clinvar |
| 665 | c.665G>A | G222E | p.Gly222Glu | Inactivating | FHH1 (heterozygous) | 122261700 | NA | SNV | Missense | Exon 4 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 666 | c.666del | I223fs* | p.Ile223fs* | Inactivating | FHH1 | 122261698 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic/Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1321376/?oq=%22NM_000388.4(CASR):c.666del%20(p.Ile223fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.666del%20(p.Ile223fs)#id_second | Clinvar |
| 668 | c.668T>C | I223T | p.Ile223Thr | Inactivating | FHH1 (heterozygous) | 122261703 | NA | SNV | Missense | Exon 4 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 668 | c.668T>C | I223T | p.Ile223Thr | Inactivating | FHH1 (heterozygous) | 122261703 | NA | SNV | Missense | Exon 4 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 674 | c.674A>C | K225T | p.Lys225Thr | Inactivating | FHH1 (heterozygous) | 122261709 | NA | SNV | Missense | Exon 4 | ECD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | NSHPT (homozygous) | 122261714 | rs1085307984 | SNV | Nonsense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Al-Khalaf | 2011 | 20972686 | 10.1007/s00431-010-1335-z | Journal article |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | NSHPT (homozygous) | 122261714 | rs1085307984 | SNV | Nonsense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Hashim | 2019 | 32120468 | 10.4038/cmj.v64i4.8988 | Journal article |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | FHH1 | 122261714 | rs1085307984 | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | FHH1 | 122261714 | rs1085307984 | SNV | Nonsense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | NSHPT (homozygous) | 122261714 | rs1085307984 | SNV | Nonsense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Padeira | 2021 | NA | 10.1159/000518849 | Conference abstract |
| 679 | c.679C>T | R227* | p.Arg227* | Inactivating | FHH1 | 122261714 | rs1085307984 | SNV | Nonsense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 679 | c.679C>G | R227G | p.Arg227Gly | NA | NA | 122261714 | rs1085307984 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/427145/?oq=%22NM_000388.4(CASR):c.679C%3EG%20(p.Arg227Gly)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.679C%3EG%20(p.Arg227Gly)#id_second | Clinvar |
| 680 | c.680G>T | R227L | p.Arg227Leu | Inactivating | NSHPT (heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Glaudo | 2016 | 27666534 | 10.1530/EJE-16-0223 | Journal article |
| 680 | c.680G>A | R227Q | p.Arg227Gln | Inactivating | FHH1 | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 680 | c.680G>A | R227Q | p.Arg227Gln | Inactivating | FHH1 (heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 680 | c.680G>A | R227Q | p.Arg227Gln | Inactivating | FHH1 | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 680 | c.680G>T | R227L | p.Arg227Leu | Inactivating | FHH1 | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 680 | c.680G>T | R227L | p.Arg227Leu | Inactivating | NSHPT (heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 680 | c.680G>T | R227L | p.Arg227Leu | Inactivating | FHH1 (Heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Veldeman | 2020 | 32306059 | 10.1007/s00223-020-00693-4 | Journal article |
| 680 | c.680G>A | R227Q | p.Arg227Gln | Inactivating | FHH1 (heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Wystrychowski | 2004 | 15572418 | 10.1210/jc.2004-1791 | Journal article |
| 680 | c.680G>A | R227Q | p.Arg227Gln | Inactivating | FHH1 (heterozygous) | 122261715 | rs28936684 | SNV | Missense | Exon 4 | ECD | Pathogenic | Chou | 1995 | 7726161 | NA | Journal article |
| 682 | c.682G>C | E228Q | p.Glu228Gln | Activating | ADH1 | 122261717 | NA | SNV | Missense | Exon 4 | ECD | NA | Conley | 2000 | 11136551 | 10.1006/mgme.2000.3096 | Journal article |
| 682 | c.682G>A | E228K | p.Glu228Lys | Activating | ADH1 | 122261717 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 682 | c.682G>A | E228K | p.Glu228Lys | Activating | ADH1 | 122261717 | NA | SNV | Missense | Exon 4 | ECD | NA | Roberts | 2019 | 31063613 | 10.1002/jbmr.3747 | Journal article |
| 682 | c.682G>A | E228K | p.Glu228Lys | Activating | ADH1 | 122261717 | NA | SNV | Missense | Exon 4 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 683 | c.683A>G | E228G | p.Glu228Gly | Activating | ADH1 | 122261718 | NA | SNV | Missense | Exon 4 | ECD | NA | Nakajima | 2009 | 19915295 | 10.2169/internalmedicine.48.2459 | Journal article |
| 683 | c.683A>C | E228A | p.Glu228Ala | Activating | ADH1 | 122261718 | NA | SNV | Missense | Exon 4 | ECD | NA | Roberts | 2019 | 31063613 | 10.1002/jbmr.3747 | Journal article |
| 704 | c.704del | I235Tfs*22 | p.Ile235Thrfs*22 | Inactivating | FHH1 | 122261739 | NA | Small deletion | Frameshift | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 706 | c.706T>G | C236G | p.Cys236Gly | Inactivating | FHH1 (heterozygous) | 122261741 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 707 | c.707_715del | C236_D238del | p.Cys236_Asp238del | Inactivating | FHH1 | 122261742 | NA | Small deletion | Deletion | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 721 | c.721G>A | E241K | p.Glu241Lys | Activating | ADH1 | 122261756 | NA | SNV | Missense | Exon 4 | ECD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 734 | c.734A>G | Q245R | p.Gln245Arg | Activating | ADH1 | 122261769 | NA | SNV | Missense | Exon 4 | ECD | NA | Conley | 2000 | 11136551 | 10.1006/mgme.2000.3096 | Journal article |
| 734 | c.734A>G | Q245R | p.Gln245Arg | Activating | ADH1 | 122261769 | NA | SNV | Missense | Exon 4 | ECD | NA | Roberts | 2019 | 31063613 | 10.1002/jbmr.3747 | Journal article |
| 734 | c.734A>G | Q245R | p.Gln245Arg | Activating | ADH1 | 122261769 | NA | SNV | Missense | Exon 4 | ECD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 741 | c.741dupT | D248* | p.Asp248* | Inactivating | FHH1 (heterozygous) | 122261776 | NA | Small insertion | Nonsense | Exon 4 | ECD | NA | Ann Tee | 2021 | 34152285 | 10.1530/EDM-21-0024 | Journal article |
| 772 | c.772_773delGTinsA | V258Rfs*47 | p.Val258Argfs*47 | Inactivating | FHH1 (heterozygous) | 122261807 | NA | Small insertion/deletion | Frameshift | Exon 4 | ECD | NA | Papadopoulou | 2016 | 27087013 | 10.4274/jcrpe.2800 | Journal article |
| 772 | c.772_773delGTinsA | V258Rfs*47 | p.Val258Argfs*47 | Inactivating | FHH1 (heterozygous) | 122261807 | NA | Small insertion/deletion | Frameshift | Exon 4 | ECD | NA | Zapanti | 2015 | 26158657 | 10.14310/horm.2002.1586 | Journal article |
| 788 | c.788C>T | T263M | p.Thr263Met | Inactivating | FHH1 (heterozygous) | 122261823 | rs201456938 | SNV | Missense | Exon 4 | ECD | VUS | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 812 | c.812C>T | S271F | p.Ser271Phe | Inactivating | FHH1 (heterozygous) | 122261847 | NA | SNV | Missense | Exon 4 | ECD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 823 | c.823_824del | D275fs* | p.Asp275fs* | NA | NA | 122261857 | rs1553766794 | Small deletion | Frameshift | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/447003/?oq=%22NM_000388.4(CASR):c.823_824del%20(p.Asp275fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.823_824del%20(p.Asp275fs)#id_first | Clinvar |
| 887 | c.887G>A | S296N | p.Ser296Asn | Inactivating | FHH1 (heterozygous) | 122261922 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 889 | c.889G>A | E297K | p.Glu297Lys | Inactivating | FHH1 (heterozygous) | 122261924 | rs121909259 | SNV | Missense | Exon 4 | ECD | Pathogenic | Brachet | 2009 | 19423559 | 10.1530/EJE-09-0257 | Journal article |
| 889 | c.889G>A | E297K | p.Glu297Lys | Inactivating | FHH1 | 122261924 | rs121909259 | SNV | Missense | Exon 4 | ECD | Pathogenic | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 889 | c.889G>A | E297K | p.Glu297Lys | Inactivating | FHH1 (heterozygous) | 122261924 | rs121909259 | SNV | Missense | Exon 4 | ECD | Pathogenic | Pollak | 1993 | 7916660 | 10.1016/0092-8674(93)90617-y | Journal article |
| 889 | c.889G>A | E297K | p.Glu297Lys | Inactivating | FHH1 (heterozygous) | 122261924 | rs121909259 | SNV | Missense | Exon 4 | ECD | Pathogenic | Woo | 2006 | 16642557 | 10.3349/ymj.2006.47.2.255 | Journal article |
| 891 | c.891G>T | E297D | p.Glu297Asp | Activating | ADH1 | 122261926 | rs1559959353 | SNV | Missense | Exon 4 | ECD | VUS | Silve | 2005 | 16147994 | 10.1074/jbc.M506263200 | Journal article |
| 893 | c.893C>T | A298V | p.Ala298Val | Inactivating | FHH1 (heterozygous) | 122261928 | rs1064797049 | SNV | Missense | Exon 4 | ECD | VUS | Lin | 2018 | NA | Lin WW, George A, Anderson RJ. A case of hypocalciuric hypercalcemia with a rare variant of casr gene mutation. Endocrine Reviews. 2018;39(2). | Conference abstract |
| 893 | c.893C>T | A298V | p.Ala298Val | Inactivating | FHH1 | 122261928 | rs1064797049 | SNV | Missense | Exon 4 | ECD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 903 | c.903del | S302fs* | p.Ser302fs* | NA | NA | 122261938 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1697584/?oq=%22NM_000388.4(CASR):c.903del%20(p.Ser302fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.903del%20(p.Ser302fs)#id_second | Clinvar |
| 905 | c.905C>T | S302F | p.Ser302Phe | Inactivating | FHH1 | 122261940 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 924 | c.924_925dup | Q309fs* | p.Gln309fs* | NA | NA | 122261957 | NA | Small insertion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1074500/?oq=%22NM_000388.4(CASR):c.924_925dup%20(p.Gln309fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.924_925dup%20(p.Gln309fs)#id_second | Clinvar |
| 961 | c.961_962del | A321fs* | p.Ala321fs* | Inactivating | FHH1 | 122261995 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1432045/?oq=%22NM_000388.4(CASR):c.961_962del%20(p.Ala321fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.961_962del%20(p.Ala321fs) | Clinvar |
| 967 | c.967A>T | K323* | p.Lys323* | Inactivating | FHH1 (heterozygous) | 122262002 | NA | SNV | Nonsense | Exon 4 | ECD | VUS | Ward | 2006 | 16649980 | 10.1111/j.1365-2265.2006.02512.x | Journal article |
| 974 | c.974G>A | G325E | p.Gly325Glu | Inactivating | FHH1 | 122262009 | rs193922444 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35803/?oq=%22NM_000388.4(CASR):c.974G%3EA%20(p.Gly325Glu)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.974G%3EA%20(p.Gly325Glu)#id_second | Clinvar |
| 995 | c.965T>C | L322P | p.Leu322Pro | Activating | ADH1 | 122262030 | NA | SNV | Missense | Exon 4 | ECD | NA | Dershem | 2020 | 32386559 | 10.1016/j.ajhg.2020.04.006 | Journal article |
| 1006 | c.1006_1008delAAG | K336del | p.Lys336del | Inactivating | FHH1 (heterozygous) | 122262041 | NA | Small deletion | Deletion | Exon 4 | ECD | NA | Warner | 2004 | 14985373 | 10.1136/jmg.2003.016725 | Journal article |
| 1015 | c.1015C>A | P339T | p.Pro339Thr | Inactivating | FHH1 (homozygous) | 122262050 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2010 | 20846291 | 10.1111/j.1365-2265.2010.03870.x | Journal article |
| 1031 | c.1031_1034 delACAAinsT | H344_N345L | p.His344_Asn345Leu | Inactivating | NSHPT (homozygous) | 122262066 | NA | Small deletion, insertion | Deletion/insertion | Exon 4 | ECD | NA | Dong | 2010 | 20631026 | 10.1210/jc.2010-0559 | Journal article |
| 1031 | c.1031_1034 delACAAinsT | H344_N345L | p.His344_Asn345Leu | Inactivating | FHH1 (heterozygous) | 122262066 | NA | Small deletion, insertion | Deletion/insertion | Exon 4 | ECD | NA | Dong | 2010 | 20631026 | 10.1210/jc.2010-0559 | Journal article |
| 1034 | c.1034A>T | N345I | p.Asn345Ile | Inactivating | FHH1 | 122262069 | rs1390562571 | SNV | Missense | Exon 4 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1040 | c.1040T>C | F347S | p.Phe347Ser | Inactivating | FHH1 | 122262075 | NA | SNV | Missense | Exon 4 | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1051 | c.1051T>G | F351V | p.Phe351Val | Inactivating | FHH1 (heterozygous) | 122262086 | NA | SNV | Missense | Exon 4 | ECD | NA | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 1054 | c.1054del | W352fs* | p.Trp352fs* | NA | NA | 122262086 | NA | Small deletion | Frameshift | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1074918/?oq=%22NM_000388.4(CASR):c.1054del%20(p.Trp352fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1054del%20(p.Trp352fs)#id_second | Clinvar |
| 1056 | c.1056G>A | W352* | p.Trp352* | Inactivating | FHH1 (heterozygous) | 122262091 | NA | SNV | Nonsense | Exon 4 | ECD | VUS/Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1081 | c.1081C>T | Q361* | p.Gln361* | NA | NA | 122262116 | NA | SNV | Nonsense | Exon 4 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1070118/?oq=%22NM_000388.4(CASR):c.1081C%3ET%20(p.Gln361Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1081C%3ET%20(p.Gln361Ter)#id_second | Clinvar |
| 1096 | c.1096G>T | G366* | p.Gly366* | Inactivating | FHH1 (heterozygous) | 122262131 | NA | SNV | Nonsense | Exon 4 | ECD | VUS | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 1096 | c.1096G>T | G366* | p.Gly366* | Inactivating | FHH1 (heterozygous) | 122262131 | NA | SNV | Nonsense | Exon 4 | ECD | VUS | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 1174 | c.1174C>T | R392* | p.Arg392* | Inactivating | NSHPT (homozygous) | 122262209 | NA | SNV | Nonsense | Exon 4 | ECD | VUS/Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1174 | c.1174C>T | R392* | p.Arg392* | Inactivating | NSHPT (homozygous) | 122262209 | NA | SNV | Nonsense | Exon 4 | ECD | VUS/Pathogenic | Ward | 2013 | 23612447 | 10.1530/EJE-13-0094 | Journal article |
| 1178 | c.1178C>G | P393R | p.Pro393Arg | Inactivating | FHH1 (heterozygous) | 122262213 | NA | SNV | Missense | Exon 4 | ECD | NA | Palmieri | 2022 | 35141253 | 10.3389/fmed.2021.809067 | Journal article |
| 1183 | c.1183T>C | C395R | p.Cys395Arg | Inactivating | FHH1 (heterozygous) | 122262218 | rs1057517712 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Forde | 2014 | 25320261 | 10.1136/bcr-2014-206473 | Journal article |
| 1183 | c.1183T>C | C395R | p.Cys395Arg | Inactivating | FHH1 | 122262218 | rs1057517712 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 1183 | c.1183T>C | C395R | p.Cys395Arg | Inactivating | FHH1 (heterozygous) | 122262218 | rs1057517712 | SNV | Missense | Exon 4 | ECD | Likely pathogenic | Vigouroux | 2000 | 10971459 | 10.1046/j.1365-2265.2000.01042.x | Journal article |
| 1189 | c.1189G>A | G397R | p.Gly397Arg | Inactivating | FHH1 (heterozygous) | 122262224 | rs1064794291 | SNV | Missense | Exon 4 | ECD | VUS | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1189 | c.1189G>A | G397R | p.Gly397Arg | Inactivating | FHH1 (heterozygous) | 122262224 | rs1064794291 | SNV | Missense | Exon 4 | ECD | VUS | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1189 | c.1189G>A | G397R | p.Gly397Arg | Inactivating | FHH1 (heterozygous) | 122262224 | rs1064794291 | SNV | Missense | Exon 4 | ECD | VUS | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1189 | c.1189G>A | G397R | p.Gly397Arg | Inactivating | FHH1 (heterozygous) | 122262224 | rs1064794291 | SNV | Missense | Exon 4 | ECD | VUS | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1199 | c.1199A>T | N400I | p.Asn400Ile | Inactivating | FHH1 (heterozygous) | 122262234 | rs576643925 | SNV | Missense | Exon 4 | ECD | VUS | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 1230 | c.1230T>A | D410E | p.Asp410Glu | Activating | ADH1 | 122262265 | NA | SNV | Missense | Exon 4 | ECD | NA | Choi | 2011 | NA | Choi B, Kim YM, Chung YS, Jung TS, Kim CH, Hong S, et al. A novel gain-of-function mutation of the calcium-sensing receptor gene, CASR D410e might be associated with autosomal dominant hypocalcemia: A series of korean hypopara registry study (2). Journal of Bone and Mineral Research. 2011;26. | Conference abstract |
| 1230 | c.1230T>A | D410E | p.Asp410Glu | Activating | ADH1 | 122262265 | NA | SNV | Missense | Exon 4 | ECD | NA | Eom | 2011 | NA | Eom YS, Choi B, Yi HS, Chung YS, Jung TS, Park SY, et al. Mutational analyses of genes in korean patients with familial or sporadic forms of isolated hypoparathyroidism: A series of korean hypopara registry study. Endocrine Reviews. 2011;32(3). | Conference abstract |
| 1230 | c.1230T>A | D410E | p.Asp410Glu | Activating | ADH1 | 122262265 | NA | SNV | Missense | Exon 4 | ECD | NA | Park | 2013 | 23009664 | 10.1111/cen.12056 | Journal article |
| 1244 | c.1244G>A | R415Q | p.Arg415Gln | Inactivating | FHH1 | 122262279 | rs193922421 | SNV | Missense | Exon 4 | ECD | Conflicting interpretations of pathogenicity | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1253 | c.1253A>G | Y418C | p.Tyr418Cys | Inactivating | FHH1 | 122262288 | NA | SNV | Missense | Exon 4 | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1256 | c.1256A>G | N419S | p.Asn419Ser | Activating | ADH1 | 122262291 | NA | SNV | Missense | Exon 4 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1258 | c.1258G>A | V420M | p.Val420Met | Inactivating | FHH1 | 122262293 | NA | SNV | Missense | Exon 4 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1263 | c.1263C>A | Y421* | p.Tyr421* | Inactivating | FHH1 (heterozygous) | 122262298 | NA | SNV | Nonsense | Exon 4 | ECD | VUS | Espejo | 2019 | NA | 10.1016/j.cca.2019.03.474 | Conference abstract |
| 1267 | c.1267_1268delinsAA | A423K | p.Ala423Lys | Inactivating | FHH1 (heterozygous) | 122262302 | NA | Small insertion/deletion | Missense | Exon 4 | ECD | NA | Livadariu | 2011 | 21566074 | 10.1530/EJE-11-0121 | Journal article |
| 1278 | c.1278delC | I427Lfs*35 | p.Ile427Leufs*35 | Inactivating | FHH1 (heterozygous) | 122262313 | NA | SNV | Frameshift | Exon 4 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 1278 | c.1278delC | I427Lfs*35 | p.Ile427Leufs*35 | Inactivating | FHH1 (heterozygous) | 122262313 | NA | SNV | Frameshift | Exon 4 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 1287 | c.1287C>A | H429Q | p.His429Gln | Inactivating | FHH1 (heterozygous) | 122262322 | rs746515147 | SNV | Missense | Exon 4 | ECD | VUS | Park | 2022 | 35586626 | 10.3389/fendo.2022.853171 | Journal article |
| 1288 | c.1288G>A | A430T | p.Ala430Thr | Inactivating | FHH1 | 122262323 | NA | SNV | Missense | Exon 4 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1289 | c.1289C>T | A430V | p.Ala430Val | Inactivating | FHH1 | 122262324 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1295 | c.1295A>G | Q432R | p.Gln432Arg | Inactivating | FHH1 | 122262330 | NA | SNV | Missense | Exon 4 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1295 | c.1295A>G | Q432R | p.Gln432Arg | Inactivating | FHH1 | 122262330 | NA | SNV | Missense | Exon 4 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1297 | c.1297G>T | D433Y | p.Asp433Tyr | Inactivating | FHH1 (heterozygous) | 122262332 | NA | SNV | Missense | Exon 4 | ECD | NA | Magno | 2020 | 32638038 | 10.1007/s00223-020-00715-1 | Journal article |
| 1342 | c.1342T>C | S448P | p.Ser448Pro | Inactivating | FHH1 (heterozygous) | 122262377 | NA | SNV | Missense | Exon 4 | ECD | NA | Dharmaraj | 2020 | 32150253 | 10.1210/clinem/dgaa111 | Journal article |
| 1342 | c.1342T>C | S448P | p.Ser448Pro | Inactivating | NSHPT (heterozygous) | 122262377 | NA | SNV | Missense | Exon 4 | ECD | NA | Dharmaraj | 2020 | 32150253 | 10.1210/clinem/dgaa111 | Journal article |
| 1370 | c.1370C>T | A457V | p.Ala457Val | Inactivating | FHH1 | 122262405 | rs1553766930 | SNV | Missense | Exon 4 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1373 | c.1373G>T | W458L | p.Trp458Leu | Inactivating | FHH1 (Heterozygous) | 122262408 | NA | SNV | Missense | Exon 4 | ECD | NA | Vishwanath | 2012 | NA | Vishwanath A, Beasley J, Krishnan S, Wierenga K. A novel calcium-sensing receptor gene (CASR) variant associated with hypocalciuric hypercalcemia. Endocrine Reviews. 2012;33(3). | Conference abstract |
| 1376 | c.1376A>G | Q459R | p.Gln459Arg | Inactivating | FHH1 (heterozygous) | 122262411 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Boisen | 2020 | 32160303 | 10.1210/clinem/dgz205 | Journal article |
| 1376 | c.1376A>G | Q459R | p.Gln459Arg | Inactivating | FHH1 (heterozygous) | 122262411 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Lietman | 2009 | 19789209 | 10.1210/jc.2008-2484 | Journal article |
| 1376 | c.1376A>G | Q459R | p.Gln459Arg | Inactivating | FHH1 | 122262411 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1376 | c.1376A>G | Q459R | p.Gln459Arg | Inactivating | FHH1 (homozygous) | 122262411 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Szczawinska | 2013 | NA | Szczawinska DB, Mayr B, Letz S, Rus R, Schnabel D, Schofl C. Identification and functional characterization of a homozygous, inactivating calcium-sensing receptor mutation in a patient with rickets. Endocrine Reviews. 2013;34(3). | Conference abstract |
| 1376 | c.1376A>G | Q459R | p.Gln459Arg | Inactivating | FHH1 (homozygous) | 122262411 | NA | SNV | Missense | Exon 4 | ECD | Pathogenic/Likely pathogenic | Szczawinska | 2014 | 24517148 | 10.1210/jc.2013-3593 | Journal article |
| 1377 | c.1377+1G>T | NA | NA | Inactivating | FHH1 (heterozygous) | 122262413 | NA | SNV | Splice donor | Exon 4-Intron 4 splice site | ECD | NA | Mahajan | 2020 | 31641801 | 10.1007/s00198-019-05170-9 | Journal article |
| 1377 | c.1377+1G>T | NA | NA | Inactivating | FHH1 | 122262413 | NA | SNV | Splice donor | Exon 4-Intron 4 splice site | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1378 | c.1378-19A>C | Q459fs*130 | p.Gln459fs*130 | Inactivating | FHH1 | 122275793 | NA | SNV | Splice acceptor/frameshift | Intron 4-Exon 5 splice site | ECD | NA | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 1378 | c.1378-2A>G | Frame shift deletion of exon 5 | Frame shift deletion of exon 5 | Inactivating | NSHPT (homozygous) | 122275810 | NA | SNV | Splice acceptor/frameshift | Intron 4-Exon 5 splice site | ECD | NA | Ahmad | 2017 | 29354167 | 10.11138/ccmbm/2017.14.3.354 | Journal article |
| 1382 | c.1382T>C | L461R | p.Leu461Arg | Inactivating | FHH1 (heterozygous) | 122275816 | NA | SNV | Missense | Exon 5 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1392 | c.1392_1404del | R465Lfs*9 | p.Arg465Leufs*9 | Inactivating | NSHPT (homozygous) | 122275826 | NA | Small deletion | Frameshift | Exon 5 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1392 | c.1392_1404del | R465Lfs*9 | p.Arg465Leufs*9 | Inactivating | NSHPT (homozygous) | 122275826 | NA | Small deletion | Frameshift | Exon 5 | ECD | NA | Soblechero | 2013 | 23817301 | 10.1159/000350540 | Journal article |
| 1393 | c.1393C>T | R465W | p.Arg465Trp | Inactivating | FHH1 (heterozygous) | 122275827 | rs751217000 | SNV | Missense | Exon 5 | ECD | Conflicting interpretations of pathogenicity | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 1393 | c.1393C>T | R465W | p.Arg465Trp | Inactivating | FHH1 | 122275827 | rs751217000 | SNV | Missense | Exon 5 | ECD | Conflicting interpretations of pathogenicity | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1394 | c.1394G>A | R465Q | p.Arg465Gln | Inactivating | FHH1 (heterozygous) | 122275828 | rs104893716 | SNV | Missense | Exon 5 | ECD | Likely pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1394 | c.1394G>A | R465Q | p.Arg465Gln | Inactivating | FHH1 (heterozygous) | 122275828 | rs104893716 | SNV | Missense | Exon 5 | ECD | Likely pathogenic | González-González | 2022 | NA | 10.4321/s1889-836x2022000200001 | Journal article |
| 1394 | c.1394G>A | R465Q | p.Arg465Gln | Inactivating | FHH1 (heterozygous) | 122275828 | rs104893716 | SNV | Missense | Exon 5 | ECD | Likely pathogenic | Leech | 2006 | 16598859 | 10.1016/j.bbrc.2006.02.018 | Journal article |
| 1394 | c.1394G>A | R465Q | p.Arg465Gln | Inactivating | FHH1 (heterozygous) | 122275828 | rs104893716 | SNV | Missense | Exon 5 | ECD | Likely pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 1394 | c.1394G>A | R465Q | p.Arg465Gln | Inactivating | FHH1 (heterozygous) | 122275828 | rs104893716 | SNV | Missense | Exon 5 | ECD | Likely pathogenic | Szalat | 2017 | 28176280 | 10.1007/s12020-017-1241-5 | Journal article |
| 1441 | c.1441G>A | E481K | p.Glu481Lys | Activating | ADH1 | 122275875 | NA | SNV | Missense | Exon 5 | ECD | NA | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 1507 | c.1507A>G | I503V | p.Ile503Val | Inactivating | FHH1 | 122275941 | rs200910001 | SNV | Missense | Exon 5 | ECD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1512 | c.1512_1515del | F505fs* | p.Phe505fs* | Inactivating | FHH1 | 122275945 | rs193922422 | Small deletion | Frameshift | Exon 5 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35777/?oq=%22NM_000388.4(CASR):c.1512_1515del%20(p.Phe505fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1512_1515del%20(p.Phe505fs)#id_second | Clinvar |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Dong | 2020 | NA | 10.3760/cma.j.cn511374-20191118-00587 | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | NSHPT (compound heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Dong | 2020 | NA | 10.3760/cma.j.cn511374-20191118-00587 | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | NSHPT (compound heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Kulkarni | 2014 | 24763815 | 10.1007/s12098-014-1442-3 | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Magno | 2020 | 32638038 | 10.1007/s00223-020-00715-1 | Journal article |
| 1525 | c.1525G>A | G509R | p.Gly509Arg | Inactivating | FHH1 (heterozygous) | 122275959 | rs193922423 | SNV | Missense | Exon 5 | ECD | Pathogenic/Likely pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1526 | c.1526G>A | G509E | p.Gly509Glu | Inactivating | FHH1 | 122275960 | rs1060502845 | SNV | Missense | Exon 5 | ECD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1542 | c.1542T>G | Y514* | p.Tyr514* | NA | NA | 122275976 | NA | SNV | Missense | Exon 5 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1453597/?oq=%22NM_000388.4(CASR):c.1542T%3EG%20(p.Tyr514Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1542T%3EG%20(p.Tyr514Ter)#id_second | Clinvar |
| 1555 | c.1555G>T | E519* | p.Glu519* | Inactivating | FHH1 (heterozygous) | 122275989 | NA | SNV | Nonsense | Exon 5 | ECD | Pathogenic | Rodrigues | 2011 | 21468522 | 10.1590/s0004-27302011000100009 | Journal article |
| 1555 | c.1555G>T | E519* | p.Glu519* | Inactivating | NSHPT (homozygous) | 122275989 | NA | SNV | Nonsense | Exon 5 | ECD | Pathogenic | Rodrigues | 2011 | 21468522 | 10.1590/s0004-27302011000100009 | Journal article |
| 1557 | c.1557_1560del | E519fs* | p.Glu519fs* | NA | NA | 122275988 | NA | Small deletion | Frameshift | Exon 5 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1376400/?oq=%22NM_000388.4(CASR):c.1557_1560del%20(p.Glu519fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1557_1560del%20(p.Glu519fs)#id_second | Clinvar |
| 1583 | c.1583T>A | I528N | p.Ile528Asn | Inactivating | FHH1 (heterozygous) | 122276017 | NA | SNV | Missense | Exon 5 | ECD | NA | Kurnaz | 2021 | NA | 10.1159/000518849 | Conference abstract |
| 1588 | c.1588T>C | W530R | p.Trp530Arg | Inactivating | FHH1 (heterozygous) | 122276022 | NA | SNV | Missense | Exon 5 | ECD | NA | Moumli | 2012 | NA | Moumli N, Zelissen PMJ, Bravenboer B. Primary hyperparathyroidism and familial hypocalciuric hypercalcemia, a diagnosis made 15 years after parathyroidectomy. Endocrine Reviews. 2012;33(3). | Conference abstract |
| 1588 | c.1588T>G | W530G | p.Trp530Gly | Inactivating | FHH1 | 122276022 | NA | SNV | Missense | Exon 5 | ECD | NA | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 1608 | c.1608G>T | E536D | p.Glu536Asp | Inactivating | FHH1 | 122276042 | NA | SNV | Missense | Exon 5 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1608 | c.1608+1G>A | NA | NA | Inactivating | NSHPT (homozygous) | 122276043 | NA | SNV | Splice donor | Exon 5-Intron 5 splice site | ECD | NA | Capozza | 2018 | 30376845 | 10.1186/s12887-018-1319-0 | Journal article |
| 1608 | c.1608+3A>G | 77-residue deletion | 77-residue deletion | Inactivating | FHH1 (heterozygous) | 122276045 | NA | SNV | Splice donor | Exon 5-Intron 5 splice site | ECD | NA | Madhavan | 2019 | 31967040 | 10.4158/ACCR-2018-0236 | Journal article |
| 1608 | c.1608+3A>C | Deletion of Exon 5 | Deletion of Exon 5 | Inactivating | FHH1 | 122276045 | NA | SNV | Splice donor | Exon 5-Intron 5 splice site | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1608 | c.1608+3A>C | Deletion of Exon 5 | Deletion of Exon 5 | Inactivating | FHH1 | 122276045 | NA | SNV | Splice donor | Exon 5-Intron 5 splice site | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1609 | c.1609-2A>G | NA | NA | NA | NA | 122282111 | rs761084315 | SNV | Splice acceptor | Exon 5-Intron 5 splice site | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/854168/?oq=%22NM_000388.4(CASR):c.1609-2A%3EG%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1609-2A%3EG#id_second | Clinvar |
| 1609 | c.1609-1G>T | Exon 6 skipping, D578fs*48 | Exon 6 skipping, p.Asp578fs*48 | Inactivating | FHH1 | 122282112 | NA | SNV | Splice acceptor/frameshift | Intron 5-Exon 6 splice site | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1609 | c.1609-1G>T | Exon 6 skipping, D578fs*48 | Exon 6 skipping, p.Asp578fs*48 | Inactivating | FHH1 | 122282112 | NA | SNV | Splice acceptor/frameshift | Intron 5-Exon 6 splice site | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1624 | c.1624T>C | C542R | p.Cys542Arg | Inactivating | FHH1 | 122282128 | NA | SNV | Missense | Exon 6 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1625 | c.1625G>A | C542Y | p.Cys542Tyr | Inactivating | FHH1 (heterozygous) | 122282129 | NA | SNV | Missense | Exon 6 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1630 | c.1630C>T | R544* | p.Arg544* | Inactivating | FHH1 (heterozygous) | 122282134 | rs886041637 | SNV | Nonsense | Exon 6 | ECD | Pathogenic | Cetani | 2019 | NA | Cetani F, Borsari S, Pardi E, Biagioni T, Bagattini B, Saponaro F, et al. Functional characterization of four mutations of the calcium sensing receptor gene identified in patients with familial hypocalciuric hypercalcemia type 1. Journal of Bone and Mineral Research. 2019;34:312. | Conference abstract |
| 1630 | c.1630C>T | R544* | p.Arg544* | Inactivating | NSHPT (compound heterozygous) | 122282134 | rs886041637 | SNV | Nonsense | Exon 6 | ECD | Pathogenic | Kulkarni | 2014 | 24763815 | 10.1007/s12098-014-1442-3 | Journal article |
| 1630 | c.1630C>T | R544* | p.Arg544* | Inactivating | FHH1 (heterozygous) | 122282134 | rs886041637 | SNV | Nonsense | Exon 6 | ECD | Pathogenic | Sorapipatcharoen | 2020 | 31883284 | 10.1111/jpc.14757 | Conference abstract |
| 1636 | c.1636T>A | C546S | p.Cys546Ser | Inactivating | FHH1 (heterozygous) | 122282140 | NA | SNV | Missense | Exon 6 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 1636 | c.1636T>A | C546S | p.Cys546Ser | Inactivating | FHH1 (heterozygous) | 122282140 | NA | SNV | Missense | Exon 6 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 1636 | c.1636T>G | C546G | p.Cys546Gly | Inactivating | FHH1 (heterozygous) | 122282140 | NA | SNV | Missense | Exon 6 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1637 | c.1637G>C | C546S | p.Cys546Ser | Inactivating | FHH1 | 122282141 | NA | SNV | Missense | Exon 6 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1645 | c.1645G>A | G549R | p.Gly549Arg | Inactivating | FHH1 (heterozygous) | 122282149 | NA | SNV | Missense | Exon 6 | ECD | NA | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 1645 | c.1645G>A | G549R | p.Gly549Arg | Inactivating | FHH1 | 122282149 | NA | SNV | Missense | Exon 6 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1645 | c.1645G>A | G549R | p.Gly549Arg | Inactivating | FHH1 | 122282149 | NA | SNV | Missense | Exon 6 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1649 | c.1649C>T | T550I | p.Thr550Ile | Inactivating | NSHPT (heterozygous) | 122282153 | NA | SNV | Missense | Exon 6 | ECD | NA | Al-Salameh | 2011 | 21566075 | 10.1530/EJE-11-0141 | Journal article |
| 1649 | c.1649C>T | T550I | p.Thr550Ile | Inactivating | FHH1 | 122282153 | NA | SNV | Missense | Exon 6 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1651 | c.1651A>G | R551G | p.Arg551Gly | Inactivating | FHH1 (heterozygous) | 122282155 | NA | SNV | Missense | Exon 6 | ECD | NA | Andrade Navarroa | 2018 | 30457731 | 10.5546/aap.2018.e757 | Journal article |
| 1651 | c.1651A>G | R551G | p.Arg551Gly | Inactivating | FHH1 (heterozygous) | 122282155 | NA | SNV | Missense | Exon 6 | ECD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1651 | c.1651A>G | R551G | p.Arg551Gly | Inactivating | FHH1 (heterozygous) | 122282155 | NA | SNV | Missense | Exon 6 | ECD | NA | Navarro | 2018 | 30457731 | 10.5546/aap.2018.e757 | Journal article |
| 1651 | c.1651A>G | R551G | p.Arg551Gly | Inactivating | FHH1 (heterozygous) | 122282155 | NA | SNV | Missense | Exon 6 | ECD | NA | Papadakis | 2016 | 28222409 | 10.14310/horm.2002.1711 | Journal article |
| 1652 | c.1652G>A | R551K | p.Arg551Lys | Inactivating | FHH1 (heterozygous) | 122282156 | rs1060502861 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Hinnie | 2009 | 20034274 | 10.1258/rsmsmj.54.4.11 | Journal article |
| 1652 | c.1652G>A | R551K | p.Arg551Lys | Inactivating | NSHPT (compound heterozygous) | 122282156 | rs1060502861 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Tõke | 2007 | 17555508 | 10.1111/j.1365-2265.2007.02896.x | Journal article |
| 1656 | c.1656delA | I554Sfs*73 | p.Ile554Serfs*73 | Inactivating | NSHPT (compound heterozygous) | 122282160 | NA | Small deletion | Frameshift | Exon 6 | ECD | NA | Panova | 2021 | 34111698 | 10.1016/j.scr.2021.102414 | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1657 | c.1657G>A | G553R | p.Gly553Arg | Inactivating | FHH1 (heterozygous) | 122282161 | rs104893719 | SNV | Missense | Exon 6 | ECD | Pathogenic | Schwarz | 2000 | 10885494 | 10.1080/003655100750044875 | Journal article |
| 1661 | c.1661T>A | I554N | p.Ile554Asn | Inactivating | FHH1 | 122282165 | NA | SNV | Missense | Exon 6 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1661 | c.1661T>A | I554N | p.Ile554Asn | Inactivating | FHH1 (heterozygous) | 122282165 | NA | SNV | Missense | Exon 6 | ECD | NA | Tsai | 2018 | NA | Tsai PH, Sung CC, Wang TH, Lin SHP. Sporadic hypocalciuric hypercalcemia caused by de novo casr mutation. Journal of the American Society of Nephrology. 2018;29:1219. | Conference abstract |
| 1661 | c.1661T>A | I554N | p.Ile554Asn | Inactivating | FHH1 | 122282165 | NA | SNV | Missense | Exon 6 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1661 | c.1661T>C | I554T | p.Ile554Thr | Activating | ADH1 | 122282165 | rs2074897929 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/992424/?oq=%22NM_000388.4(CASR):c.1661T%3EC%20(p.Ile554Thr)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1661T%3EC%20(p.Ile554Thr)#id_second | Clinvar |
| 1663 | c.1663A>G | I555V | p.Ile555Val | Inactivating | FHH1 (heterozygous) | 122282167 | rs777646067 | SNV | Missense | Exon 6 | ECD | VUS | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1664 | c.1664T>C | I555T | p.Ile555Thr | Inactivating | FHH1 (heterozygous) | 122282168 | rs1576875819 | SNV | Missense | Exon 6 | ECD | Conflicting interpretations of pathogenicity | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1664 | c.1664T>C | I555T | p.Ile555Thr | Inactivating | FHH1 | 122282168 | rs1576875819 | SNV | Missense | Exon 6 | ECD | Conflicting interpretations of pathogenicity | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1664 | c.1664T>C | I555T | p.Ile555Thr | Inactivating | NSHPT (heterozygous) | 122282168 | rs1576875819 | SNV | Missense | Exon 6 | ECD | Conflicting interpretations of pathogenicity | Tonyushkina | 2012 | 22620673 | 10.1186/1687-9856-2012-13 | Journal article |
| 1664 | c.1664T>C | I555T | p.Ile555Thr | Inactivating | FHH1 (heterozygous) | 122282168 | rs1576875819 | SNV | Missense | Exon 6 | ECD | Conflicting interpretations of pathogenicity | Wang | 2020 | 32871939 | 10.1097/MD.0000000000021940 | Journal article |
| 1666 | c.1666G>A | E556K | p.Glu556Lys | Activating | ADH1 | 122282170 | NA | SNV | Missense | Exon 6 | ECD | NA | Livadariu | 2011 | 21566074 | 10.1530/EJE-11-0121 | Journal article |
| 1666 | c.1666G>A | E556K | p.Glu556Lys | Activating | ADH1 | 122282170 | NA | SNV | Missense | Exon 6 | ECD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 1670 | c.1670G>A | G557E | p.Gly557Glu | Inactivating | FHH1 | 122282174 | rs1576875835 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1670 | c.1670G>A | G557E | p.Gly557Glu | Inactivating | FHH1 | 122282174 | rs1576875835 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Nakamura | 2001 | 11762699 | 10.1385/ENDO:15:3:277 | Journal article |
| 1670 | c.1670G>A | G557E | p.Gly557Glu | Inactivating | FHH1 (Heterozygous) | 122282174 | rs1576875835 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Zajíčková | 2020 | 33094630 | 10.33549/physiolres.934522 | Journal article |
| 1672 | c.1672dup | E558fs* | p.Glu558fs* | Inactivating | FHH1 | 122282171 | NA | Small insertion | Frameshift | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1678560/?oq=%22NM_000388.4(CASR):c.1672dup%20(p.Glu558fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1672dup%20(p.Glu558fs)#id_second | Clinvar |
| 1673 | c.1673A>G | E558G | p.Glu558Gly | NA | NA | 122282177 | rs1553768726 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/432667/?oq=%22NM_000388.4(CASR):c.1673A%3EG%20(p.Glu558Gly)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1673A%3EG%20(p.Glu558Gly)#id_second | Clinvar |
| 1676 | c.1676C>A | P559H | p.Pro559His | Inactivating | FHH1 | 122282180 | rs193922425 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35780/?oq=%22NM_000388.4(CASR):c.1676C%3EA%20(p.Pro559His)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1676C%3EA%20(p.Pro559His) | Clinvar |
| 1682 | c.1682G>C | C561S | p.Cys561Ser | Inactivating | FHH1 | 122282186 | NA | SNV | Missense | Exon 6 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1685 | c.1685G>A | C562Y | p.Cys562Tyr | Inactivating | FHH1 (heterozygous) | 122282189 | NA | SNV | Missense | Exon 6 | ECD | NA | Burski | 2002 | 11889154 | 10.1210/jcem.87.3.8304 | Journal article |
| 1685 | c.1685G>A | C562Y | p.Cys562Tyr | Inactivating | FHH1 (heterozygous) | 122282189 | NA | SNV | Missense | Exon 6 | ECD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1685 | c.1685G>A | C562Y | p.Cys562Tyr | Inactivating | FHH1 | 122282189 | NA | SNV | Missense | Exon 6 | ECD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1685 | c.1685_1686delinsCT | C562S | p.Cys562Ser | NA | NA | 122282189 | rs193922427 | Small insertion/deletion | Missense | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35782/?oq=%22NM_000388.4(CASR):c.1685_1686delinsCT%20(p.Cys562Ser)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1685_1686delinsCT%20(p.Cys562Ser)#id_first | Clinvar |
| 1685 | c.1685G>C | C562S | p.Cys562Ser | Inactivating | FHH1 | 122282189 | rs193922426 | SNV | Missense | Exon 6 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35781/?oq=%22NM_000388.4(CASR):c.1685G%3EC%20(p.Cys562Ser)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1685G%3EC%20(p.Cys562Ser)#id_second | Clinvar |
| 1693 | c.1693T>G | C565G | p.Cys565Gly | Inactivating | FHH1 | 122282197 | rs1559967708 | SNV | Missense | Exon 6 | ECD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1697 | c.1697_1698delTG | V566Gfs*4 | p.Val566Glyfs*4 | Inactivating | FHH1 | 122282201 | NA | Small deletion | Frameshift | Exon 6 | ECD | NA | Cordes | 2016 | 27926951 | 10.1055/s-0042-120415 | Journal article |
| 1703 | c.1703G>A | C568Y | p.Cys568Tyr | Inactivating | FHH1 (heterozygous) | 122282207 | NA | SNV | Missense | Exon 6 | ECD | VUS | Gunganah | 2014 | 25045523 | 10.1530/EDM-14-0050 | Journal article |
| 1703 | c.1703G>A | C568Y | p.Cys568Tyr | Inactivating | FHH1 | 122282207 | NA | SNV | Missense | Exon 6 | ECD | VUS | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 1706 | c.1706C>A | P569H | p.Pro569His | Activating | ADH1 | 122282210 | NA | SNV | Missense | Exon 6 | ECD | NA | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 1706 | c.1706C>A | P569H | p.Pro569His | Activating | ADH1 | 122282210 | NA | SNV | Missense | Exon 6 | ECD | NA | Nakamura | 2013 | 23966241 | 10.1210/jc.2013-1974 | Journal article |
| 1709 | c.1709A>G | D570G | p.Asp570Gly | Inactivating | FHH1 | 122282213 | NA | SNV | Missense | Exon 6 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1711 | c.1711G>T | G571W | p.Gly571Trp | Inactivating | FHH1 (heterozygous) | 122282215 | NA | SNV | Missense | Exon 6 | ECD | NA | Kim | 2015 | 26386835 | 10.1007/s00774-015-0713-z | Journal article |
| 1718 | c.1718A>G | Y573C | p.Tyr573Cys | Inactivating | FHH1 (heterozygous) | 122282222 | NA | SNV | Missense | Exon 6 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1719 | c.1719T>A | Y573* | p.Tyr573* | Inactivating | FHH1 (heterozygous) | 122282223 | NA | SNV | Nonsense | Exon 6 | ECD | NA | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1719 | c.1719T>A | Y573* | p.Tyr573* | Inactivating | FHH1 (heterozygous) | 122282223 | NA | SNV | Nonsense | Exon 6 | ECD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1719 | c.1719T>A | Y573* | p.Tyr573* | Inactivating | FHH1 (heterozygous) | 122282223 | NA | SNV | Nonsense | Exon 6 | ECD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1733 | c.1733-255_2450del | Shorter protein due to affected exon 7 splice site | Shorter protein due to affected exon 7 splice site | Inactivating | FHH1 (heterozygous) | 122283432 | NA | CNV | Deletion | Intron 6-Exon 7 | ECD | NA | García-Castaño | 2018 | 30530875 | 10.1530/EDM-18-0114 | Journal article |
| 1740 | c.1740T>A | S580R | p.Ser580Arg | Inactivating | FHH1 (heterozygous) | 122283694 | NA | SNV | Missense | Exon 7 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 1740 | c.1740T>A | S580R | p.Ser580Arg | Inactivating | FHH1 (heterozygous) | 122283694 | NA | SNV | Missense | Exon 7 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 1744 | c.1744T>C | C582R | p.Cys582Arg | Inactivating | FHH1 | 122283698 | NA | SNV | Missense | Exon 7 | ECD | NA | Joseph | 2022 | NA | https://jme.bioscientifica.com/abstract/journals/jme/69/1/JME-21-0263.xml | Journal article |
| 1744 | c.1744T>A | C582S | p.Cys582Ser | NA | NA | 122283698 | rs2074920676 | SNV | Missense | Exon 7 | ECD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/853442/?oq=%22NM_000388.4(CASR):c.1744T%3EA%20(p.Cys582Ser)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1744T%3EA%20(p.Cys582Ser) | Clinvar |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1745 | c.1745G>T | C582F | p.Cys582Phe | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1745 | c.1745G>T | C582F | p.Cys582Phe | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1745 | c.1745G>T | C582F | p.Cys582Phe | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1745 | c.1745G>T | C582F | p.Cys582Phe | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | NSHPT (compound heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Leunbach | 2021 | 33748353 | 10.1016/j.bonr.2021.100761 | Journal article |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1745 | c.1745G>T | C582F | p.Cys582Phe | Inactivating | FHH1 (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | NSHPT (heterozygous) | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 1745 | c.1745G>A | C582Y | p.Cys582Tyr | Inactivating | FHH1 | 122283699 | rs104893690 | SNV | Missense | Exon 7 | ECD | Pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1746 | c.1746_1747insT | N583* | p.Asn583* | Inactivating | FHH1 | 122283700 | NA | Small insertion | Nonsense | Exon 7 | ECD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1746 | c.1746_1747insT | N583* | p.Asn583* | Inactivating | FHH1 | 122283700 | NA | Small insertion | Nonsense | Exon 7 | ECD | VUS | Pidasheva | 2006 | 16740594 | 10.1093/hmg/ddl145 | Journal article |
| 1750 | c.1750A>T | K584* | p.Lys584* | Inactivating | FHH1 (heterozygous) | 122283704 | rs1057523748 | SNV | Missense | Exon 7 | ECD | VUS/Likely pathogenic | Hans | 2022 | NA | Hans SK, Levine SN. A previously unreported mutation of the calcium sensing receptor causing familial hypocalciuric hypercalcemia. Endocrine Reviews. 2017;38(3) | Conference abstract |
| 1754 | c.1754G>T | C585F | p.Cys585Phe | Inactivating | FHH1 | 122283708 | NA | SNV | Missense | Exon 7 | ECD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1754 | c.1754G>T | C585F | p.Cys585Phe | Inactivating | FHH1 | 122283708 | NA | SNV | Missense | Exon 7 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1759 | c.1759dup | D587fs* | p.Asp587fs* | NA | NA | 122283712 | NA | Small insertion | Frameshift | Exon 7 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1373420/?oq=%22NM_000388.4(CASR):c.1759dup%20(p.Asp587fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1759dup%20(p.Asp587fs) | Clinvar |
| 1772 | c.1772C>G | S591C | p.Ser591Cys | Inactivating | FHH1 | 122283726 | NA | SNV | Missense | Exon 7 | ECD | NA | Nyweide | 2006 | 17110864 | 10.1097/01.pec.0000238747.19477.d3 | Journal article |
| 1772 | c.1772C>G | S591C | p.Ser591Cys | Inactivating | FHH1 | 122283726 | NA | SNV | Missense | Exon 7 | ECD | NA | Taki | 2015 | 26019872 | 10.1530/EDM-15-0016 | Journal article |
| 1781 | c.1781A>C | N594T | p.Asn594Thr | Inactivating | FHH1 | 122283735 | NA | SNV | Missense | Exon 7 | ECD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1783 | c.1783del | H595fs* | p.His595fs* | NA | NA | 122283736 | NA | Small deletion | Frameshift | Exon 7 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1071398/?oq=%22NM_000388.4(CASR):c.1783del%20(p.His595fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1783del%20(p.His595fs)#id_second | Clinvar |
| 1782 | c.1782C>T | H595Y | p.His595Tyr | Inactivating | FHH1 (heterozygous) | 122283736 | NA | SNV | Missense | Exon 7 | ECD | NA | Cetani | 2009 | 19073830 | 10.1530/EJE-08-0798 | Journal article |
| 1801 | c.1801A>C | K601Q | p.Lys601Gln | Inactivating | FHH1 (heterozygous) | 122283755 | NA | SNV | Missense | Exon 7 | ECD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 1801 | c.1801A>C | K601Q | p.Lys601Gln | Inactivating | FHH1 (heterozygous) | 122283755 | NA | SNV | Missense | Exon 7 | ECD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 1802 | c.1802del | K601fs* | p.Lys601fs* | NA | NA | 122283755 | NA | Small deletion | Frameshift | Exon 7 | ECD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1425273/?oq=%22NM_000388.4(CASR):c.1802del%20(p.Lys601fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1802del%20(p.Lys601fs)#id_second | Clinvar |
| 1803 | c.1803G>C | K601N | p.Lys601Asn | Inactivating | FHH1 | 122283757 | NA | SNV | Missense | Exon 7 | ECD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Álvarez-Hernández | 2003 | 14519094 | 10.1677/jme.0.0310255 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Iglesias | 2006 | 17043292 | 10.1136/jnnp.2006.097162 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Schmidt | 2018 | 30276462 | 10.1007/s00423-018-1711-0 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Tan | 2003 | 12574188 | 10.1210/jc.2002-020081 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1810 | c.1810G>A | E604K | p.Glu604Lys | Activating | ADH1 | 122283764 | rs104893712 | SNV | Missense | Exon 7 | ECD | Pathogenic | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 1817 | c.1817T>C | L606P | p.Leu606Pro | Inactivating | NSHPT (homozygous) | 122283771 | NA | SNV | Missense | Exon 7 | ECD | VUS | Wejaphikul | 2023 | 36626889 | 10.1159/000528568 | Journal article |
| 1820 | c.1820C>A | S607* | p.Ser607* | Inactivating | FHH1 (heterozygous) | 122283774 | NA | SNV | Nonsense | Exon 7 | ECD | VUS | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 1829 | c.1829A>G | E610G | p.Glu610Gly | Activating | ADH1 | 122283783 | NA | SNV | Missense | Exon 7 | ECD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1835 | c.1835C>T | F612S | p.Phe612Ser | Activating | ADH1 | 122283789 | rs104893698 | SNV | Missense | Exon 7 | ECD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 1835 | c.1835C>T | F612S | p.Phe612Ser | Activating | ADH1 | 122283789 | rs104893698 | SNV | Missense | Exon 7 | ECD | Pathogenic | Pearce | 1996 | 8813042 | 10.1056/NEJM199610103351505 | Journal article |
| 1836 | c.1836G>A | G613E | p.Gly613Glu | Inactivating | NSHPT (homozygous) | 122283790 | NA | SNV | Missense | Exon 7 | TMD | NA | Gulcan-Kersin | 2020 | 33147586 | 10.1159/000510623 | Journal article |
| 1836 | c.1836delT | F612fs*15 | p.Phe612fs*15 | Inactivating | FHH1 (heterozygous) | 122283790 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1837 | c.1837G>C | G613R | p.Gly613Arg | Inactivating | FHH1 (heterozygous) | 122283791 | rs1060502842 | SNV | Missense | Exon 7 | TMD | VUS/Likely pathogenic | Rasmussen | 2011 | 22142470 | 10.1186/1752-1947-5-564 | Journal article |
| 1846 | c.1846C>G | L616V | p.Leu616Val | Activating | ADH1 | 122283800 | rs104893703 | SNV | Missense | Exon 7 | TMD | Pathogenic | Stock | 1999 | 10487661 | 10.1210/jcem.84.9.5977 | Journal article |
| 1849 | c.1849del | T627fs* | p.Thr627fs* | NA | NA | 122283803 | rs1553768938 | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/446987/?oq=%22NM_000388.4(CASR):c.1849del%20(p.Thr617fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1849del%20(p.Thr617fs) | Clinvar |
| 1852 | c.1852del | L618fs* | p.Leu618fs* | NA | NA | 122283804 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1449397/?oq=%22NM_000388.4(CASR):c.1852del%20(p.Leu618fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1852del%20(p.Leu618fs) | Clinvar |
| 1855 | c.1855_1857delTT | L618fs*81 | p.Leu618fs*81 | Inactivating | FHH1 | 122283809 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous, neonatal) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Braun | 2010 | PMC7102049 | 10.1007/s00467-010-1577-z | Conference abstract |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous, neonatal) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Braun | 2010 | 22001932 | 10.1159/000321348 | Conference abstract |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1868 | c.1868G>A | G623D | p.Gly623Asp | Inactivating | FHH1 (heterozygous) | 122283822 | NA | SNV | Missense | Exon 7 | TMD | VUS | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 1868 | c.1868del | G623fs* | p.Gly623fs* | NA | NA | 122283820 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1076659/?oq=%22NM_000388.4(CASR):c.1868del%20(p.Gly623fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1868del%20(p.Gly623fs)#id_second | Clinvar |
| 1884 | c.1884del | F629fs* | p.Phe629fs* | Inactivating | FHH1 | 122283837 | rs193922429 | Small deletion | Frameshift | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35784/?oq=%22NM_000388.4(CASR):c.1884del%20(p.Phe629fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1884del%20(p.Phe629fs)#id_second | Clinvar |
| 1898 | c.1898_1932dup | A645fs* | p.Ala645fs* | NA | NA | 122283850 | rs2074924614 | Small insertion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/950326/?oq=%22NM_000388.4(CASR):c.1898_1932dup%20(p.Ala645fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1898_1932dup%20(p.Ala645fs) | Clinvar |
| 1901 | c.1901T>C | F634S | p.Phe634Ser | Inactivating | FHH1 | 122283855 | NA | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1901 | c.1901T>C | F634S | p.Phe634Ser | Inactivating | FHH1 | 122283855 | NA | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1913 | c.1913G>T | R638L | p.Arg638Leu | Inactivating | FHH1 (heterozygous) | 122283867 | rs201852643 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Corrado | 2015 | 25828954 | 10.1002/jbmr.2516 | Journal article |
| 1913 | c.1913G>T | R638L | p.Arg638Leu | Inactivating | NSHPT (compound heterozygous) | 122283867 | rs201852643 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Corrado | 2015 | 25828954 | 10.1002/jbmr.2516 | Journal article |
| 1913 | c.1913dup | N639fs* | p.Asn639fs* | NA | NA | 122283866 | rs1576877163 | Small insertion | Frameshift | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/804661/?oq=%22NM_000388.4(CASR):c.1913dup%20(p.Asn639fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1913dup%20(p.Asn639fs)#id_second | Clinvar |
| 1919 | c.1919C>T | T640I | p.Thr640Ile | Inactivating | FHH1 | 122283873 | rs1559968450 | SNV | Missense | Exon 7 | TMD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1919 | c.1919C>T | T640I | p.Thr640Ile | Inactivating | FHH1 | 122283873 | rs1559968450 | SNV | Missense | Exon 7 | TMD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1929 | c.1929_1930delGT | F644Yfs*73 | p.Phe644Tyrfs*73 | Inactivating | FHH1 (heterozygous) | 122283883 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 1934 | c.1934C>A | A645D | p.Ala645Asp | Inactivating | FHH1 (heterozygous) | 122283888 | rs193922430 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | NSHPT (compound heterozygous) | 122283896 | rs104893705 | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Edouard | 2013 | NA | Edouard T, Mouly C, Mimoun E, Gennero I, Magdelaine C, Salles JP. Management of a new case of neonatal hypocalciuric hypercalcaemia related to mutation of the calcium-sensing receptor gene with bone abnormalities. Hormone Research in Paediatrics. 2013;80:223. | Conference abstract |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | FHH1 (heterozygous) | 122283896 | rs104893705 | SNV | Missense | Exon 7 | TMD | VUS/Pathogenic | Jap | 2001 | 11231970 | 10.1210/jcem.86.1.7149 | Journal article |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | FHH1 | 122283896 | rs104893705 | SNV | Missense | Exon 7 | TMD | VUS/Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | FHH1 | 122283896 | rs104893705 | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | NSHPT (compound heterozygous) | 122283896 | rs104893705 | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Ward | 2004 | 15292296 | 10.1210/jc.2003-031653 | Journal article |
| 1942 | c.1942C>T | R648* | p.Arg648* | Inactivating | FHH1 (heterozygous) | 122283896 | rs104893705 | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Yamauchi | 2002 | 12469911 | 10.1359/jbmr.2002.17.12.2174 | Journal article |
| 1949 | c.1949T>C | L650P | p.Leu650Pro | Inactivating | FHH1 (heterozygous) | 122283903 | NA | SNV | Missense | Exon 7 | TMD | NA | Warner | 2004 | 14985373 | 10.1136/jmg.2003.016725 | Journal article |
| 1952 | c.1952_1966del | S651_L655del | p.Ser651_Leu655del | Inactivating | FHH1 (heterozygous) | 122283906 | NA | SNV | Missense | Exon 7 | TMD | NA | Monastiri | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 1952 | c.1952_1966del | S651_L655del | p.Ser651_Leu655del | Inactivating | FHH1 (heterozygous) | 122283906 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Sfar | 2012 | 21667241 | 10.1007/s11033-011-0990-0 | Journal article |
| 1952 | c.1952_1966del | S651_L655del | p.Ser651_Leu655del | Inactivating | NSHPT (homozygous) | 122283906 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Sfar | 2012 | 21667241 | 10.1007/s11033-011-0990-0 | Journal article |
| 1963 | c.1963_1965delCTC | L655del | p.Leu655del | Inactivating | FHH1 | 122283917 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 1963 | c.1963_1965delCTC | L655del | p.Leu655del | Inactivating | FHH1 | 122283917 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1970 | c.1970C>G | S657C | p.Ser657Cys | Activating | ADH1 | 122283924 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 1970 | c.1970C>G | S657C | p.Ser657Cys | Activating | ADH1 | 122283924 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 1972 | c.1972del | L658fs* | p.Leu658fs* | NA | NA | 122283924 | rs1553768972 | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/463917/?oq=%22NM_000388.4(CASR):c.1972del%20(p.Leu658fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.1972del%20(p.Leu658fs)#id_second | Clinvar |
| 1982 | c.1982G>A | C661Y | p.Cys661Tyr | Inactivating | FHH1 | 122283936 | NA | SNV | Missense | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 1983 | c.1983C>G | C661W | p.Cys661Trp | Inactivating | FHH1 | 122283937 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 1997 | c.1997_1998insT | L656fs*42 | p.Leu656fs*42 | Inactivating | FHH1 (heterozygous) | 122283951 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 1997 | c.1997_1998insT | L656fs*42 | p.Leu656fs*42 | Inactivating | FHH1 (heterozygous) | 122283951 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 1997 | c.1997_1998insT | L656fs*42 | p.Leu656fs*42 | Inactivating | FHH1 (heterozygous) | 122283951 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 1997 | c.1997_1998insT | L656fs*42 | p.Leu656fs*42 | Inactivating | FHH1 (heterozygous) | 122283951 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 (heterozygous) | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 (heterozygous) | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 (heterozygous) | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 2008 | c.2009G>A | G670E | p.Gly670Glu | Inactivating | NSHPT (compound heterozygous) | 122283963 | rs104893700 | SNV | Missense | Exon 7 | TMD | Pathogenic | Kobayashi | 1997 | 9253359 | 10.1210/jcem.82.8.4135 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 (heterozygous) | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2008 | c.2008G>A | G670R | p.Gly670Arg | Inactivating | FHH1 (heterozygous) | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 2008 | c.2008G>C | G670R | p.Gly670Arg | NA | NA | 122283962 | NA | SNV | Missense | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1397805/?oq=%22NM_000388.4(CASR):c.2008G%3EC%20(p.Gly670Arg)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2008G%3EC%20(p.Gly670Arg)#id_second | Clinvar |
| 2010 | c.2010_2011del | E681fs* | p.Glu681fs* | NA | NA | 122283962 | rs2074927630 | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/945014/?oq=%22NM_000388.4(CASR):c.2010_2011del%20(p.Glu671fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2010_2011del%20(p.Glu671fs)#id_second | Clinvar |
| 2013 | c.2013G>C | E671D | p.Glu671Asp | Inactivating | FHH1 (homozygous) | 122283967 | NA | SNV | Missense | Exon 7 | TMD | NA | Borsari | 2017 | 28620806 | 10.1007/s40618-017-0710-2 | Journal article |
| 2013 | c.2013G>C | E671D | p.Glu671Asp | Inactivating | FHH1 (heterozygous) | 122283967 | NA | SNV | Missense | Exon 7 | TMD | NA | Cetani | 2019 | NA | Cetani F, Borsari S, Pardi E, Biagioni T, Bagattini B, Saponaro F, et al. Functional characterization of four mutations of the calcium sensing receptor gene identified in patients with familial hypocalciuric hypercalcemia type 1. Journal of Bone and Mineral Research. 2019;34:312. | Conference abstract |
| 2014 | c.2014C>A | P672T | p.Pro672Thr | Inactivating | FHH1 | 122283968 | rs193922431 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35786/?oq=%22NM_000388.4(CASR):c.2014C%3EA%20(p.Pro672Thr)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2014C%3EA%20(p.Pro672Thr)#id_second | Clinvar |
| 2024 | c.2024G>A | W675* | p.Trp675* | Inactivating | FHH1 | 122283978 | NA | SNV | Nonsense | Exon 7 | TMD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2029 | c.2029dup | C677Lfs*31 | p.Cys677Leufs*31 | Inactivating | FHH1 | 122283983 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2029 | c.2029dup | C677Lfs*31 | p.Cys677Leufs*31 | Inactivating | FHH1 | 122283983 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2030 | c.2030G>A | C677Y | p.Cys677Tyr | Inactivating | FHH1 (heterozygous) | 122283984 | NA | SNV | Missense | Exon 7 | TMD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2030 | c.2030del | C677fs* | p.Cys677fs* | Inactivating | FHH1 | 122283984 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1377560/?oq=%22NM_000388.4(CASR):c.2030del%20(p.Cys677fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2030del%20(p.Cys677fs)#id_first | Clinvar |
| 2033 | c.2033G>A | R678H | p.Arg678His | Inactivating | FHH1 | 122283987 | rs1278025825 | SNV | Missense | Exon 7 | TMD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2036 | c.2036T>C | L679P | p.Leu679Pro | Inactivating | FHH1 | 122283990 | rs1553768983 | SNV | Missense | Exon 7 | TMD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2038 | c.2038C>G | R680G | p.Arg680Gly | Activating | ADH1 | 122283992 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | NSHPT (compound heterozygous) | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | NSHPT (homozygous) | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Hassan | 2021 | 34887979 | 10.11604/pamj.2021.40.105.29527 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | FHH1 | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2038 | c.2038C>A | R680S | p.Arg680Ser | Inactivating | FHH1 | 122283992 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | FHH1 | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | FHH1 | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | FHH1 (heterozygous) | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 2038 | c.2038C>T | R680C | p.Arg680Cys | Inactivating | NSHPT (compound heterozygous) | 122283992 | rs767363250 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Waller | 2004 | 15241688 | 10.1007/s00431-004-1491-0 | Journal article |
| 2039 | c.2039G>A | R680H | p.Arg680His | Inactivating | NSHPT (homozygous) | 122283993 | rs773146939 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Haider | 2022 | NA | 10.1159/000518849 | Conference abstract |
| 2039 | c.2039G>A | R680H | p.Arg680His | Inactivating | FHH1 (heterozygous) | 122283993 | rs773146939 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Kurian | 2021 | 34993031 | 10.7759/cureus.20057 | Journal article |
| 2039 | c.2039G>A | R680H | p.Arg680His | Inactivating | FHH1 | 122283993 | rs773146939 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2039 | c.2039G>A | R680H | p.Arg680His | Inactivating | FHH1 | 122283993 | rs773146939 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2039 | c.2039G>A | R680H | p.Arg680His | Inactivating | FHH1 | 122283993 | rs773146939 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2042 | c.2042A>G | Q681R | p.Gln681Arg | Activating | ADH1 | 122283996 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Guarnieri | 2012 | 22789683 | 10.1016/j.ymgme.2012.06.012 | Journal article |
| 2042 | c.2042A>G | Q681R | p.Gln681Arg | Activating | ADH1 | 122283996 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Mazurek | 2022 | NA | 10.1016/j.eprac.2022.03.234 | Journal article |
| 2043 | c.2043G>T | Q681H | p.Gln681His | Activating | ADH1 | 122283997 | rs121909261 | SNV | Missense | Exon 7 | TMD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2043 | c.2043G>T | Q681H | p.Gln681His | Activating | ADH1 | 122283997 | rs121909261 | SNV | Missense | Exon 7 | TMD | Pathogenic | Baron | 1996 | 8733126 | 10.1093/hmg/5.5.601 | Journal article |
| 2045 | c.2045C>T | P682L | p.Pro682Leu | Inactivating | NSHPT (compound heterozygous) | 122283999 | rs1553768989 | SNV | Missense | Exon 7 | TMD | VUS | Leunbach | 2021 | 33748353 | 10.1016/j.bonr.2021.100761 | Journal article |
| 2048 | c.2048C>A | A683D | p.Ala683Asp | Inactivating | FHH1 | 122284002 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2054 | c.2054G>T | G685V | p.Gly685Val | Inactivating | NSHPT (homozygous) | 122284008 | NA | SNV | Missense | Exon 7 | TMD | NA | Alqadi | 2018 | 32289793 | 10.1159/000492307 | Conference abstract |
| 2054 | c.2054G>C | G685A | p.Gly685Ala | Inactivating | FHH1 (heterozygous) | 122284008 | rs1576877377 | SNV | Missense | Exon 7 | TMD | VUS | Saba | 2017 | NA | 10.1159/000481424 | Conference abstract |
| 2065 | c.2065G>A | V689M | p.Val689Met | Inactivating | FHH1 (heterozygous) | 122284019 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2065 | c.2065G>A | V689M | p.Val689Met | Inactivating | FHH1 (heterozygous) | 122284019 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Sharma | 2020 | 32843465 | 10.1136/bcr-2020-237036 | Journal article |
| 2065 | c.2065G>A | V689M | p.Val689Met | Inactivating | FHH1 (heterozygous) | 122284019 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Warner | 2004 | 14985373 | 10.1136/jmg.2003.016725 | Journal article |
| 2069 | c.2069T>G | L690R | p.Leu690Arg | Inactivating | FHH1 | 122284023 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2072 | c.2072G>A | C691Y | p.Cys691Tyr | Inactivating | FHH1 | 122284026 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2078 | c.2078C>T | S693L | p.Ser693Leu | Inactivating | FHH1 | 122284032 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2083 | c.2083_2106dup | I695_V702dup | p.Ile695_Val702dup | Inactivating | FHH1 | 122284037 | NA | Small insertion | Small insertion | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2086 | c.2086C>G | L696V | p.Leu696Val | Activating | ADH1 | 122284040 | NA | SNV | Missense | Exon 7 | TMD | NA | Gomes | 2020 | 32513763 | 10.1136/bcr-2020-234391 | Journal article |
| 2086 | c.2086C>G | L696V | p.Leu696Val | Activating | ADH1 | 122284040 | NA | SNV | Missense | Exon 7 | TMD | NA | Silvestre | 2018 | NA | Silvestre CF, Carvalho MR, Bugalho MJ. Familial hypoparathyroidism: A novel casr gene mutation. Endocrine Reviews. 2018;39(2). | Conference abstract |
| 2089 | c.2089G>A | V697M | p.Val697Met | Inactivating | FHH1 (heterozygous) | 122284043 | NA | SNV | Missense | Exon 7 | TMD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2089 | c.2089G>A | V697M | p.Val697Met | Inactivating | FHH1 (heterozygous) | 122284043 | NA | SNV | Missense | Exon 7 | TMD | NA | López | 2012 | 21643651 | 10.1007/s00431-011-1504-8 | Journal article |
| 2089 | c.2089G>A | V697M | p.Val697Met | Inactivating | FHH1 | 122284043 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2096 | c.2096C>A | T699N | p.Thr699Asn | Inactivating | FHH1 (heterozygous) | 122284050 | NA | SNV | Missense | Exon 7 | TMD | NA | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 2096 | c.2096C>A | T699N | p.Thr699Asn | Inactivating | FHH1 (heterozygous) | 122284050 | NA | SNV | Missense | Exon 7 | TMD | NA | Mullin | 2022 | 35356007 | 10.1210/jendso/bvac025 | Journal article |
| 2097 | c.2097del | N700fs* | p.Asn700fs* | NA | NA | 122284050 | rs1576877427 | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/804662/?oq=%22NM_000388.4(CASR):c.2097del%20(p.Asn700fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2097del%20(p.Asn700fs)#id_second | Clinvar |
| 2101 | c.2101delC | R701fs* | p.Arg701fs* | Inactivating | FHH1 (heterozygous) | 122284055 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2101 | c.2101C>G | R701G | p.Arg701Gly | Inactivating | FHH1 (heterozygous) | 122284055 | NA | SNV | Missense | Exon 7 | TMD | NA | Mullin | 2022 | 35356007 | 10.1210/jendso/bvac025 | Journal article |
| 2102 | c.2102G>C | R701P | p.Arg701Pro | Inactivating | FHH1 (heterozygous) | 122284056 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2120 | c.2120A>T | E707V | p.Glu707Val | Inactivating | FHH1 (heterozygous) | 122284074 | NA | SNV | Missense | Exon 7 | TMD | NA | Quaglia | 2012 | NA | 10.1093/ndt/gfs201 | Conference abstract |
| 2120 | c.2120A>T | E707V | p.Glu707Val | Inactivating | FHH1 (heterozygous) | 122284074 | NA | SNV | Missense | Exon 7 | TMD | NA | Strata | 2014 | 25104082 | 10.1093/ndt/gfu065 | Journal article |
| 2146 | c.2146C>T | R716C | p.Arg716Cys | Inactivating | FHH1 (heterozygous) | 122284100 | NA | SNV | Missense | Exon 7 | TMD | NA | Ghazvani | 2016 | 27957351 | 10.1155/2016/2725486 | Journal article |
| 2148 | c.2148dup | K717fs* | p.Lys717fs* | NA | NA | 122284101 | NA | Small insertion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1442327/?oq=%22NM_000388.4(CASR):c.2148dup%20(p.Lys717fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2148dup%20(p.Lys717fs)#id_second | Clinvar |
| 2154 | c.2154G>A | W718* | p.Trp718* | Inactivating | FHH1 (heterozygous) | 122284108 | NA | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Frank-Raue | 2011 | 21521328 | 10.1111/j.1365-2265.2011.04059.x | Journal article |
| 2154 | c.2154G>A | W718* | p.Trp718* | Inactivating | FHH1 | 122284108 | NA | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2154 | c.2154G>A | W718* | p.Trp718* | Inactivating | FHH1 | 122284108 | NA | SNV | Nonsense | Exon 7 | TMD | VUS/Pathogenic | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 2154 | c.2154delinsCC | W718fs* | p.Trp718fs* | NA | NA | 122284108 | rs1064793353 | Small insertion/deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/418664/?oq=%22NM_000388.4(CASR):c.2154delinsCC%20(p.Trp718fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2154delinsCC%20(p.Trp718fs)#id_second | Clinvar |
| 2156 | c.2156G>A | W719* | p.Trp719* | Inactivating | FHH1 | 122284110 | NA | SNV | Nonsense | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1683780/?oq=%22NM_000388.4(CASR):c.2156G%3EA%20(p.Trp719Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2156G%3EA%20(p.Trp719Ter)#id_second | Clinvar |
| 2165 | c.2165A>G | N722S | p.Asn722Ser | Activating | ADH1 | 122284119 | NA | SNV | Missense | Exon 7 | TMD | NA | Schoutteten | 2017 | 28741586 | NA | Journal article |
| 2165 | c.2165del | N722fs* | p.Asn722fs* | NA | NA | 122284118 | rs1559968729 | Small deletion | Frameshift | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/562459/?oq=%22NM_000388.4(CASR):c.2165del%20(p.Asn722fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2165del%20(p.Asn722fs) | Clinvar |
| 2168 | c.2168T>A | L723Q | p.Leu723Gln | Activating | ADH1 in mouse | 122284122 | NA | SNV | Missense | Exon 7 | TMD | NA | Hough | 2004 | 15347804 | 10.1073/pnas.0405516101 | Journal article |
| 2180 | c.2180T>A | L727Q | p.Leu727Gln | Activating | ADH1 | 122284134 | rs104893718 | SNV | Missense | Exon 7 | TMD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2180 | c.2180T>A | L727Q | p.Leu727Gln | Activating | ADH1 | 122284134 | rs104893718 | SNV | Missense | Exon 7 | TMD | Pathogenic | Mittleman | 2006 | 16608894 | 10.1210/jc.2005-2605 | Journal article |
| 2182 | c.2182G>A | V728F | p.Val728Phe | Inactivating | FHH1 (heterozygous) | 122284136 | NA | SNV | Missense | Exon 7 | TMD | NA | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 2182 | c.2182G>A | V728F | p.Val728Phe | Inactivating | FHH1 (heterozygous) | 122284136 | NA | SNV | Missense | Exon 7 | TMD | NA | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 2182 | c.2182G>T | V728F | p.Val728Phe | Inactivating | FHH1 (heterozygous) | 122284136 | NA | SNV | Missense | Exon 7 | TMD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 2182 | c.2182G>A | V728F | p.Val728Phe | Inactivating | FHH1 (heterozygous) | 122284136 | NA | SNV | Missense | Exon 7 | TMD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2201 | c.2201T>G | M734R | p.Met734Arg | Inactivating | FHH1 | 122284155 | NA | SNV | Missense | Exon 7 | TMD | NA | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2201 | c.2201T>G | M734R | p.Met734Arg | Inactivating | FHH1 | 122284155 | NA | SNV | Missense | Exon 7 | TMD | NA | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 2201 | c.2201T>C | M734T | p.Met734Thr | Activating | ADH1 | 122284155 | NA | SNV | Missense | Exon 7 | TMD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 2203 | c.2203C>A | Q735K | p.Gln735Lys | Inactivating | FHH1 | 122284157 | rs1553769052 | SNV | Missense | Exon 7 | TMD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2204 | c.2204A>C | Q735P | p.Gln735Pro | Activating | ADH1 | 122284158 | NA | SNV | Missense | Exon 7 | TMD | NA | Wong | 2011 | 21471599 | https://www.hkmj.org/abstracts/v17n2/157.htm | Journal article |
| 2217 | c.2217T>A | C739* | p.Cys739* | Inactivating | NSHPT (compound heterozygous) | 122284171 | NA | SNV | Missense | Exon 7 | TMD | VUS | Panova | 2021 | 34111698 | 10.1016/j.scr.2021.102414 | Journal article |
| 2224 | c.2224T>C | W742R | p.Trp742Arg | Inactivating | FHH1 (heterozygous) | 122284178 | NA | SNV | Missense | Exon 7 | TMD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 2224 | c.2224T>C | W742R | p.Trp742Arg | Inactivating | FHH1 (heterozygous) | 122284178 | NA | SNV | Missense | Exon 7 | TMD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2228 | c.2228T>C | L743P | p.Leu743Pro | Inactivating | FHH1 | 122284182 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2237 | c.2237C>T | A746P | p.Ala746Pro | Inactivating | FHH1 | 122284191 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2239 | c.2239_2240delinsT | P747fs*28 | p.Pro747fs*28 | Inactivating | NSHPT (homozygous) | 122284193 | NA | Small deletoin/insertion | Missense | Exon 7 | TMD | NA | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 2240 | c.2240C>T | P747L | p.Pro747Leu | Inactivating | FHH1 | 122284194 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2242 | c.2242C>T | P748S | p.Pro748Ser | Inactivating | FHH1 (heterozygous) | 122284196 | rs2074934184 | SNV | Missense | Exon 7 | TMD | VUS | Cetani | 2009 | 19073830 | 10.1530/EJE-08-0798 | Journal article |
| 2243 | c.2243C>G | P748R | p.Pro748Arg | Inactivating | FHH1 (heterozygous) | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Arshad | 2021 | 32892159 | 10.1136/postgradmedj-2020-137718 | Journal article |
| 2243 | c.2243C>G | P748R | p.Pro748Arg | Inactivating | FHH1 (heterozygous) | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2243 | c.2243C>T | P748L | p.Pro748Leu | Inactivating | FHH1 (heterozygous) | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2243 | c.2243C>G | P748R | p.Pro748Arg | Inactivating | FHH1 | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2243 | c.2243C>A | P748H | p.Pro748His | Inactivating | FHH1 | 122284197 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2243 | c.2243C>G | P748R | p.Pro748Arg | Inactivating | FHH1 | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2243 | c.2243C>G | P748R | p.Pro748Arg | Inactivating | FHH1 | 122284197 | rs193922433 | SNV | Missense | Exon 7 | TMD | Conflicting interpretations of pathogenicity | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2244 | c.2244delinsC | S749fs* | p.Ser749fs* | NA | NA | 122284198 | rs1553769059 | Small insertion/deletion | Frameshift | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/snp/rs1553769059/#clinical_significance | Clinvar |
| 2254 | c.2254C>T | R752C | p.Arg752Cys | Inactivating | FHH1 (heterozygous) | 122284208 | rs193922434 | SNV | Missense | Exon 7 | TMD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2254 | c.2254del | R752fs* | p.Arg752fs* | NA | NA | 122284207 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1068041/?oq=%22NM_000388.4(CASR):c.2254del%20(p.Arg752fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2254del%20(p.Arg752fs)#id_second | Clinvar |
| 2269 | c.2269G>A | E757K | p.Glu757Lys | Activating | ADH1 | 122284223 | rs2074935149 | SNV | Missense | Exon 7 | TMD | VUS | Gomes | 2020 | 32513763 | 10.1136/bcr-2020-234391 | Journal article |
| 2269 | c.2269G>A | E757K | p.Glu757Lys | Activating | ADH1 | 122284223 | rs2074935149 | SNV | Missense | Exon 7 | TMD | VUS | Kwan | 2018 | 30306783 | 10.1530/EDM-18-0107 | Journal article |
| 2279 | c.2279T>A | I760N | p.Ile760Asn | Inactivating | FHH1 (heterozygous) | 122284233 | NA | SNV | Missense | Exon 7 | TMD | NA | Wu | 2017 | 28690912 | 10.1038/boneres.2017.1 | Journal article |
| 2281 | c.2281_2283delATG | I761del | p.Ile761del | Inactivating | FHH1 | 122284235 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2282 | c.2282T>A | I761N | p.Ile761Asn | Inactivating | FHH1 (heterozygous) | 122284236 | NA | SNV | Missense | Exon 7 | TMD | NA | Bhangu | 2018 | 30554440 | 10.1002/hed.25568 | Journal article |
| 2282 | c.2282T>A | I761N | p.Ile761Asn | Inactivating | FHH1 (heterozygous) | 122284236 | NA | SNV | Missense | Exon 7 | TMD | NA | Bhangu | 2022 | 35242665 | 10.21037/gs-21-577 | Journal article |
| 2293 | c.2293T>C | C765R | p.Cys765Arg | Inactivating | FHH1 (heterozygous) | 122284247 | rs1363412937 | SNV | Missense | Exon 7 | TMD | VUS | Dixit | 2011 | 21369680 | 10.2340/00015555-1054 | Journal article |
| 2295 | c.2295C>G | C765W | p.Cys765Trp | Inactivating | FHH1 (heterozygous) | 122284249 | NA | SNV | Missense | Exon 7 | TMD | VUS | Cetani | 2009 | 19073830 | 10.1530/EJE-08-0798 | Journal article |
| 2297 | c.2297_2298dup | E767fs* | p.Glu767fs* | NA | NA | 122284249 | NA | Small insertion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1068229/?oq=%22NM_000388.4(CASR):c.2297_2298dup%20(p.Glu767fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2297_2298dup%20(p.Glu767fs) | Clinvar |
| 2299 | c.2299G>A | E767K | p.Glu767Lys | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Gómez-Martín | 2023 | 36609127 | 10.1016/j.ando.2022.12.001 | Journal article |
| 2299 | c.2299G>A | E767K | p.Glu767Lys | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2299 | c.2299G>C | E767K | p.Glu767Lys | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2299 | c.2299G>A | E767K | p.Glu767Lys | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2299 | c.2299G>C | E767Q | p.Glu767Gln | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Letz | 2010 | 20668040 | 10.1210/jc.2010-0651 | Journal article |
| 2299 | c.2299G>A | E767K | p.Glu767Lys | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Uçkun-Kitapçi | 2005 | 15551332 | 10.1002/ajmg.a.30403 | Journal article |
| 2299 | c.2299G>C | E767Q | p.Glu767Gln | Activating | ADH1 | 122284253 | NA | SNV | Missense | Exon 7 | TMD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 2303 | c.2303G>T | G768V | p.Gly768Val | Inactivating | NSHPT (homozygous) | 122284257 | rs201858689 | SNV | Missense | Exon 7 | TMD | VUS | Diaz-Thomas | 2011 | 24854525 | 10.1515/jpem-2013-0343 | Journal article |
| 2303 | c.2303G>T | G768V | p.Gly768Val | Inactivating | NSHPT (homozygous) | 122284257 | rs201858689 | SNV | Missense | Exon 7 | TMD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2318 | c.2318T>G | L773R | p.Leu773Arg | Activating | ADH1 | 122284272 | rs104893699 | SNV | Missense | Exon 7 | TMD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2319 | c.2319delG | L773fs* | p.Leu773fs* | Inactivating | NSHPT (homozygous) | 122284273 | NA | SNV | Missense | Exon 7 | TMD | NA | Sadacharan | 2020 | 32699790 | 10.4103/ijem.IJEM_53_20 | Journal article |
| 2320 | c.2320G>A | G774S | p.Gly774Ser | Inactivating | FHH1 (heterozygous) | 122284274 | NA | SNV | Missense | Exon 7 | TMD | NA | Quaglia | 2012 | NA | 10.1093/ndt/gfs201 | Conference abstract |
| 2320 | c.2320G>A | G774S | p.Gly774Ser | Inactivating | FHH1 (heterozygous) | 122284274 | NA | SNV | Missense | Exon 7 | TMD | NA | Strata | 2014 | 25104082 | 10.1093/ndt/gfu065 | Journal article |
| 2331 | c.2331_2332insAGC | I777_G778insS | p.Ile777_Gly778insSer | Inactivating | FHH1 | 122284285 | NA | Small inserion | Insertion | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2332 | c.2332G>C | G778R | p.Gly778Arg | Inactivating | FHH1 (heterozygous) | 122284286 | rs1479933693 | SNV | Missense | Exon 7 | TMD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2332 | c.2332G>C | G778R | p.Gly778Arg | Inactivating | FHH1 | 122284286 | rs1479933693 | SNV | Missense | Exon 7 | TMD | VUS | Khairi | 2020 | 32761341 | 10.1007/s12672-020-00394-2 | Journal article |
| 2332 | c.2332G>C | G778R | p.Gly778Arg | Inactivating | FHH1 | 122284286 | rs1479933693 | SNV | Missense | Exon 7 | TMD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2333 | c.2333G>A | G778D | p.Gly778Asp | Inactivating | FHH1 | 122284287 | rs2074936560 | SNV | Missense | Exon 7 | TMD | VUS | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2333 | c.2333G>A | G778D | p.Gly778Asp | Inactivating | FHH1 (heterozygous) | 122284287 | rs2074936560 | SNV | Missense | Exon 7 | TMD | VUS | Ward | 2006 | 16649980 | 10.1111/j.1365-2265.2006.02512.x | Journal article |
| 2335 | c.2335T>C | Y779H | p.Tyr779His | Inactivating | FHH1 | 122284289 | NA | SNV | Missense | Exon 7 | TMD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2351 | c.2351C>T | A784V | p.Ala784Val | Activating | ADH1 | 122284305 | NA | SNV | Missense | Exon 7 | TMD | NA | van Megan | 2022 | 35313217 | 10.1016/j.ebiom.2022.103947 | Journal article |
| 2351 | c.2351C>T | A784V | p.Ala784Val | Activating | ADH1 | 122284305 | NA | SNV | Missense | Exon 7 | TMD | NA | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 2362 | c.2362T>C | F788L | p.Phe788Leu | Inactivating | FHH1 (heterozygous) | 122284316 | rs104893711 | SNV | Missense | Exon 7 | TMD | Pathogenic | Hendy | 2023 | 12915654 | 10.1210/jc.2003-030409 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Abbas | 2022 | NA | 10.1016/j.eprac.2022.03.235 | Conference abstract |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Elston | 2022 | 35402765 | 10.1210/jendso/bvac042 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Kamiyoshi | 2015 | 26323216 | 10.1007/s10157-015-1160-9 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Mora | 2005 | 16333828 | 10.1002/ajmg.a.31054 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Rossi | 2019 | 31763346 | 10.1177/2329048X19876199 | Journal article |
| 2363 | c.2363T>G | F788C | p.Phe788Cys | Activating | ADH1 | 122284317 | rs104893701 | SNV | Missense | Exon 7 | TMD | Pathogenic | Watanabe | 1998 | 9661634 | 10.1210/jcem.83.7.4920 | Journal article |
| 2364 | c.2364delC | Y789fs* | p.Tyr789fs* | Inactivating | NSHPT (homozygous) | 122284318 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Gupta | 2022 | 35380381 | 10.1007/s12098-022-04169-1 | Journal article |
| 2367 | c.2367_2369del | F789del | p.Phe789del | Inactivating | FHH1 (heterozygous) | 122284321 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Ubetagoyena Arrieta | 2011 | NA | 10.1016/j.anpedi.2010.10.001 | Journal article |
| 2371 | c.2371G>C | A791P | p.Ala791Pro | Inactivating | FHH1 (heterozygous) | 122284325 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2383 | c.2383delC | R795Gfs*42 | p.Arg795Gfs*42 | Inactivating | FHH1 (heterozygous) | 122284337 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 2383 | c.2383delC | R795Gfs*42 | p.Arg795Gfs*42 | Inactivating | FHH1 (heterozygous) | 122284337 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 (heterozygous) | 122284337 | rs121909258 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 | 122284337 | rs121909258 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 2383 | c.2383delC | R795Gfs*42 | p.Arg795Gfs*42 | Inactivating | FHH1 (heterozygous) | 122284337 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 2383 | c.2383delC | R795Gfs*42 | p.Arg795Gfs*42 | Inactivating | FHH1 (heterozygous) | 122284337 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 (heterozygous) | 122284337 | rs121909258 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Konijnenberg | 2012 | NA | 10.1007/s00467-012-2232-7 | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 (heterozygous) | 122284337 | NA | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2383 | c.2383delC | R795Gfs*42 | p.Arg795Gfs*42 | Inactivating | FHH1 (heterozygous) | 122284337 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 (heterozygous) | 122284337 | rs121909258 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Pollak | 1993 | 7916660 | 10.1016/0092-8674(93)90617-y | Journal article |
| 2383 | c.2383C>T | R795W | p.Arg795Trp | Inactivating | FHH1 | 122284337 | rs121909258 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2384 | c.2384G>A | R795Q | p.Arg795Gln | Inactivating | FHH1 | 122284338 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2384 | c.2384G>C | R795P | p.Arg795Pro | Inactivating | FHH1 | 122284338 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2386 | c.2386A>T | K796* | p.Lys796* | Inactivating | FHH1 | 122284340 | NA | SNV | Nonsense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2386 | c.2386A>T | K796* | p.Lys796* | Inactivating | FHH1 | 122284340 | NA | SNV | Nonsense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2392 | c.2392C>A | P798T | p.Pro798Thr | Inactivating | FHH1 | 122284346 | NA | SNV | Missense | Exon 7 | TMD | NA | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 2392 | c.2392C>A | P798T | p.Pro798Thr | Inactivating | FHH1 (heterozygous) | 122284346 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2392 | c.2392C>A | P798T | p.Pro798Thr | Inactivating | FHH1 (heterozygous) | 122284346 | NA | SNV | Missense | Exon 7 | TMD | NA | Lam | 2005 | 15963484 | 10.1016/j.cccn.2005.04.026 | Journal article |
| 2393 | c.2393C>T | P798L | p.Pro798Leu | Inactivating | FHH1 (heterozygous) | 122284347 | rs1060502856 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2393 | c.2393C>T | P798L | p.Pro798Leu | Inactivating | FHH1 (heterozygous) | 122284347 | rs1060502856 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Udelsman | 2015 | NA | 10.1002/jbmr.2763 | Journal article |
| 2395 | c.2395G>A | E799L | p.Glu799Leu | Activating | ADH1 | 122284349 | NA | SNV | Missense | Exon 7 | TMD | NA | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 2399 | c.2399A>C | N800T | p.Asn800Thr | Activating | ADH1 | 122284353 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2401 | c.2401T>C | F801L | p.Phe801Leu | Inactivating | FHH1 | 122284355 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2401 | c.2401T>C | F801L | p.Phe801Leu | Inactivating | FHH1 | 122284355 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2404 | c.2404A>C | N802H | p.Asn802His | NA | NA | 122284358 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1338619/?oq=%22NM_000388.4(CASR):c.2404A%3EC%20(p.Asn802His)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2404A%3EC%20(p.Asn802His) | Clinvar |
| 2405 | c.2405A>G | N802S | p.Asn802Ser | Inactivating | FHH1 (heterozygous) | 122284359 | rs140022350 | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Lia-Baldini | 2013 | 23169696 | 10.1530/EJE-12-0714 | Journal article |
| 2405 | c.2405A>T | N802I | p.Asn802Ile | Activating | ADH1 | 122284359 | NA | SNV | Missense | Exon 7 | TMD | NA | Lia-Baldini | 2013 | 23169696 | 10.1530/EJE-12-0714 | Journal article |
| 2405 | c.2405A>T | N802I | p.Asn802Ile | Activating | ADH1 | 122284359 | NA | SNV | Missense | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2405 | c.2405A>T | N802I | p.Asn802Ile | Activating | ADH1 | 122284359 | NA | SNV | Missense | Exon 7 | TMD | NA | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 2410 | c.2410G>A | A804T | p.Ala804Thr | Inactivating | FHH1 | 122284364 | rs777219967 | SNV | Missense | Exon 7 | TMD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2411 | c.2411C>A | A804D | p.Ala804Asp | Inactivating | FHH1 | 122284365 | NA | SNV | Missense | Exon 7 | TMD | NA | Grant | 2012 | 23077345 | 10.1210/me.2012-1232 | Journal article |
| 2411 | c.2411C>A | A804D | p.Ala804Asp | Inactivating | FHH1 | 122284365 | NA | SNV | Missense | Exon 7 | TMD | NA | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2411 | c.2411C>A | A804D | p.Ala804Asp | Inactivating | FHH1 (heterozygous) | 122284365 | NA | SNV | Missense | Exon 7 | TMD | NA | Miyashiro | 2004 | 15579196 | 10.1111/j.1365-2265.2004.02163.x | Journal article |
| 2414 | c.2414A>G | K805R | p.Lys805Arg | Inactivating | FHH1 (heterozygous) | 122284368 | NA | SNV | Missense | Exon 7 | TMD | NA | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2414 | c.2414A>G | K805R | p.Lys805Arg | Inactivating | FHH1 (heterozygous) | 122284368 | NA | SNV | Missense | Exon 7 | TMD | NA | Sagi | 2020 | 33434173 | 10.1530/EDM-20-0084 | Journal article |
| 2415 | c.2415del | K805fs* | p.Lys805fs* | NA | NA | 122284369 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1066835/?oq=%22NM_000388.4(CASR):c.2415del%20(p.Lys805fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2415del%20(p.Lys805fs)#id_second | Clinvar |
| 2417 | c.2417T>C | F806S | p.Phe806Ser | Activating | ADH1 | 122284371 | rs104893693 | SNV | Missense | Exon 7 | TMD | Pathogenic | Theman | 2009 | 19063686 | 10.1359/JBMR.081233 | Journal article |
| 2417 | c.2417T>C | F806S | p.Phe806Ser | Activating | ADH1 | 122284371 | rs104893693 | SNV | Missense | Exon 7 | TMD | Pathogenic | Baron | 1996 | 8733126 | 10.1093/hmg/5.5.601 | Journal article |
| 2417 | c.2417T>C | F806S | p.Phe806Ser | Activating | ADH1 | 122284371 | rs104893693 | SNV | Missense | Exon 7 | TMD | Pathogenic | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 2420 | c.2420T>A | I807N | p.Ile807Asn | Inactivating | FHH1 | 122284374 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2422 | c.2422A>C | T808P | p.Thr808Pro | Inactivating | FHH1 (heterozygous) | 122284376 | NA | SNV | Missense | Exon 7 | TMD | NA | Mullin | 2022 | 35356007 | 10.1210/jendso/bvac025 | Journal article |
| 2423 | c.2393C>T | P798L | p.Pro798Leu | Inactivating | FHH1 (heterozygous) | 122284347 | NA | SNV | Missense | Exon 7 | TMD | Pathogenic/Likely pathogenic | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2425 | c.2425T>A | F809L | p.Phe809Leu | Inactivating | FHH1 | 122284379 | rs2074938402 | SNV | Missense | Exon 7 | TMD | VUS | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 2425 | c.2425T>C | F809L | p.Phe809Leu | Inactivating | FHH1 | 122284379 | rs2074938402 | SNV | Missense | Exon 7 | TMD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2425 | c.2425T>A | F809L | p.Phe809Leu | Inactivating | FHH1 (heterozygous) | 122284379 | rs2074938402 | SNV | Missense | Exon 7 | TMD | VUS | Timmers | 2006 | 16882283 | 10.1111/j.1365-2796.2006.01684.x | Journal article |
| 2425 | c.2425T>A | F809I | p.Phe809Ile | Inactivating | FHH1 | 122284379 | rs2074938402 | SNV | Missense | Exon 7 | TMD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2427 | c.2427C>G | F809L | p.Phe809Leu | Inactivating | FHH1 | 122284381 | rs2074938402 | SNV | Missense | Exon 7 | TMD | VUS | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2429 | c.2429G>A | S810N | p.Ser810Asn | Activating | ADH1 | 122284383 | rs2074938472 | SNV | Missense | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/978027/?oq=%22NM_000388.4(CASR):c.2429G%3EA%20(p.Ser810Asn)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2429G%3EA%20(p.Ser810Asn)#id_second | Clinvar |
| 2431 | c.2431A>G | M811V | p.Met811Val | Activating | ADH1 | 122284385 | rs1057521129 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Schouten | 2011 | 21441391 | 10.1258/acb.2010.010139 | Journal article |
| 2435 | c.2435T>G | L812R | p.Leu812Arg | Inactivating | FHH1 | 122284389 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2435 | c.2435T>C | L812P | p.Leu812Pro | Inactivating | FHH1 | 122284389 | rs193922435 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35791/?oq=%22NM_000388.4(CASR):c.2435T%3EC%20(p.Leu812Pro)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2435T%3EC%20(p.Leu812Pro)#id_second | Clinvar |
| 2443 | c.2443_2445delTTC | F815del | p.Phe815del | Activating | ADH1 | 122284397 | NA | Small deletion | Deletion | Exon 7 | TMD | NA | Gagliardi | 2016 | 27177819 | 10.1111/cen.13104 | Journal article |
| 2446 | c.2446A>G | I816V | p.Ile816Val | Inactivating | FHH1 (heterozygous) | 122284400 | NA | SNV | Missense | Exon 7 | TMD | VUS | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2446 | c.2446A>G | I816V | p.Ile816Val | Inactivating | FHH1 (heterozygous) | 122284400 | NA | SNV | Missense | Exon 7 | TMD | VUS | Jones | 2020 | 32537548 | 10.1002/jbm4.10362 | Journal article |
| 2446 | c.2446A>G | I816V | p.Ile816Val | Inactivating | FHH1 (heterozygous) | 122284400 | NA | SNV | Missense | Exon 7 | TMD | VUS | Mogas | 2018 | 29899992 | 10.1530/EDM-18-0009 | Journal article |
| 2447 | c.2447T>C | I816T | p.Ile816Thr | Inactivating | FHH1 (heterozygous) | 122284401 | NA | SNV | Missense | Exon 7 | TMD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2447 | c.2447T>C | I816T | p.Ile816Thr | Inactivating | FHH1 (heterozygous) | 122284401 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2449 | c.2449G>A | V817I | p.Val817Ile | Inactivating | FHH1 | 122284403 | rs1057518933 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2449 | c.2449G>A | V817I | p.Val817Ile | Inactivating | FHH1 (heterozygous) | 122284403 | rs1057518933 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Pearce | 1995 | 8675635 | 10.1172/JCI118335 | Journal article |
| 2453 | c.2453G>T | W818L | p.Trp818Leu | Activating | ADH1 | 122284407 | NA | SNV | Missense | Exon 7 | TMD | NA | Schouten | 2011 | 21441391 | 10.1258/acb.2010.010139 | Journal article |
| 2454 | c.2454G>A | W828* | p.Trp828* | NA | NA | 122284408 | rs1576878011 | SNV | Nonsense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/804665/?oq=%22NM_000388.4(CASR):c.2454G%3EA%20(p.Trp818Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2454G%3EA%20(p.Trp818Ter)#id_second | Clinvar |
| 2458 | c.2458T>G | S820A | p.Ser820Ala | Inactivating | FHH1 (heterozygous) | 122284412 | rs2074939020 | SNV | Missense | Exon 7 | TMD | VUS | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 2458 | c.2458T>G | S820A | p.Ser820Ala | Inactivating | FHH1 | 122284412 | rs2074939020 | SNV | Missense | Exon 7 | TMD | VUS | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 2459 | c.2459C>T | S820F | p.Ser820Phe | Activating | ADH1 | 122284413 | rs104893710 | SNV | Missense | Exon 7 | TMD | Pathogenic | Belli | 2019 | NA | 10.1159/000501868 | Conference abstract |
| 2459 | c.2459C>T | S820F | p.Ser820Phe | Activating | ADH1 | 122284413 | rs104893710 | SNV | Missense | Exon 7 | TMD | Pathogenic | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2459 | c.2459C>T | S820F | p.Ser820Phe | Activating | ADH1 | 122284413 | rs104893710 | SNV | Missense | Exon 7 | TMD | Pathogenic | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 2459 | c.2459C>A | S820Y | p.Ser820Tyr | Activating | ADH1 | 122284413 | NA | SNV | Missense | Exon 7 | TMD | NA | Hawkes | 2020 | 33112267 | 10.1530/EJE-20-0710 | Journal article |
| 2459 | c.2459C>T | S820F | p.Ser820Phe | Activating | ADH1 | 122284413 | rs104893710 | SNV | Missense | Exon 7 | TMD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2459 | c.2459C>T | S820F | p.Ser820Phe | Activating | ADH1 | 122284413 | rs104893710 | SNV | Missense | Exon 7 | TMD | Pathogenic | Nagase | 2002 | 12050233 | 10.1210/jcem.87.6.8531 | Journal article |
| 2461 | c.2461T>C | F821L | p.Phe821Leu | Activating | ADH1 | 122284415 | NA | SNV | Missense | Exon 7 | TMD | NA | Gevers | 2017 | NA | Gevers EF, Buck J, Ashman N, Thakker RV, Allgrove J. Continuous subcutaneous PTH infusion in autosomal dominant hypocalcaemia. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 2461 | c.2461T>C | F821L | p.Phe821Leu | Activating | ADH1 | 122284415 | NA | SNV | Missense | Exon 7 | TMD | NA | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2461 | c.2461T>C | F821L | p.Phe821Leu | Activating | ADH1 | 122284415 | NA | SNV | Missense | Exon 7 | TMD | NA | Sastre | 2021 | 34233101 | 10.1056/NEJMc2034981 | Journal article |
| 2461 | c.2461T>C | F821L | p.Phe821Leu | Activating | ADH1 | 122284415 | NA | SNV | Missense | Exon 7 | TMD | NA | Shiohara | 2004 | 14677060 | 10.1007/s00431-003-1331-7 | Journal article |
| 2461 | c.2461T>C | F821L | p.Phe821Leu | Activating | ADH1 | 122284415 | NA | SNV | Missense | Exon 7 | TMD | NA | Shiohara | 2006 | 16983178 | 10.1507/endocrj.k06-053 | Journal article |
| 2467 | c.2467C>T | P823S | p.Pro823Ser | Inactivating | FHH1 | 122284421 | rs1576878027 | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2470 | c.2470G>C | A824P | p.Ala824Pro | Activating | ADH1 | 122284424 | NA | SNV | Missense | Exon 7 | TMD | NA | Chapman | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 2470 | c.2470G>T | A824S | p.Ala824Ser | Activating | ADH1 | 122284424 | NA | SNV | Missense | Exon 7 | TMD | NA | Chapman | 2010 | NA | 10.1159/000321348 | Conference abstract |
| 2470 | c.2470G>C | A824P | p.Ala824Pro | Activating | ADH1 | 122284424 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2474 | c.2474A>T | Y825F | p.Tyr825Phe | Activating | ADH1 | 122284428 | NA | SNV | Missense | Exon 7 | TMD | NA | Moon | 2018 | 29969884 | 10.6065/apem.2018.23.2.107 | Journal article |
| 2479 | c.2479A>G | S827G | p.Ser827Gly | Inactivating | FHH1 | 122284433 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2479 | c.2479A>C | S827R | p.Ser827Arg | Inactivating | FHH1 (heterozygous) | 122284433 | NA | SNV | Missense | Exon 7 | TMD | NA | Szalat | 2017 | 28176280 | 10.1007/s12020-017-1241-5 | Journal article |
| 2479 | c.2479A>C | S827R | p.Ser827Arg | Inactivating | FHH1 | 122284433 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2482 | c.2482A>C | T828P | p.Thr828Pro | Inactivating | FHH1 | 122284436 | rs794729230 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/202205/?oq=%22NM_000388.4(CASR):c.2482A%3EC%20(p.Thr828Pro)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2482A%3EC%20(p.Thr828Pro)#id_first | Clinvar |
| 2483 | c.2483C>A | T828D | p.Thr828Asp | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Gevers | 2017 | NA | Gevers EF, Buck J, Ashman N, Thakker RV, Allgrove J. Continuous subcutaneous PTH infusion in autosomal dominant hypocalcaemia. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 2483 | c.2483C>A | T828D | p.Thr828Asp | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Giner | 2013 | NA | 10.1055/s-0033-1337875 | Journal article |
| 2483 | c.2483C>A | T828D | p.Thr828Asp | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 2483 | c.2483C>T | T828I | p.Thr828Ile | Inactivating | FHH1 (heterozygous) | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 29463778 | 10.1126/scisignal.aan3714 | Journal article |
| 2483 | c.2483C>A | T828N | p.Thr828Asn | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2483 | c.2483C>A | T828N | p.Thr828Asn | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Sastre | 2021 | 34233101 | 10.1056/NEJMc2034981 | Journal article |
| 2483 | c.2483C>A | T828N | p.Thr828Asn | Activating | ADH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 2483 | c.2483C>T | T828I | p.Thr828Ile | Inactivating | FHH1 | 122284437 | NA | SNV | Missense | Exon 7 | TMD | NA | Gorvin | 2018 | 30052933 | 10.1093/hmg/ddy263 | Journal article |
| 2486 | c.2486A>G | Y829C | p.Tyr829Cys | Activating | ADH1 | 122284440 | NA | SNV | Missense | Exon 7 | TMD | NA | Choi | 2015 | 25932037 | 10.3345/kjp.2015.58.4.148 | Journal article |
| 2486 | c.2486A>G | Y829C | p.Tyr829Cys | Activating | ADH1 with Bartter syndrome (heterozygous) | 122284440 | NA | SNV | Missense | Exon 7 | TMD | NA | Choi | 2015 | 25932037 | 10.3345/kjp.2015.58.4.148 | Journal article |
| 2486 | c.2486A>G | Y829C | p.Tyr829Cys | Activating | ADH1 | 122284440 | NA | SNV | Missense | Exon 7 | TMD | NA | Gevers | 2017 | NA | Gevers EF, Buck J, Ashman N, Thakker RV, Allgrove J. Continuous subcutaneous PTH infusion in autosomal dominant hypocalcaemia. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 2486 | c.2486A>G | Y829C | p.Tyr829Cys | Activating | ADH1 | 122284440 | NA | SNV | Missense | Exon 7 | TMD | NA | Sastre | 2021 | 34233101 | 10.1056/NEJMc2034981 | Journal article |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | Harari-Shaham | 2019 | NA | 10.1038/s41431-018-0248-6 | Conference abstract |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | Letz | 2010 | 20668040 | 10.1210/jc.2010-0651 | Journal article |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | McDonald | 2019 | NA | 10.1007/s11845-019-02073-w | Conference abstract |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2488 | c.2488G>A | G830S | p.Gly830Ser | Activating | ADH1 | 122284442 | NA | SNV | Missense | Exon 7 | TMD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 2489 | c.2489G>A | G830D | p.Gly830Asp | Inactivating | FHH1 | 122284443 | rs193922436 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35792/?oq=%22NM_000388.4(CASR):c.2489G%3EA%20(p.Gly830Asp)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2489G%3EA%20(p.Gly830Asp)#id_second | Clinvar |
| 2492 | c.2492A>C | K831T | p.Lys831Thr | Activating | ADH1 | 122284446 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2494 | c.2494T>C | F832L | p.Phe832Leu | Activating | ADH1 | 122284448 | NA | SNV | Missense | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Hawkes | 2020 | 33112267 | 10.1530/EJE-20-0710 | Journal article |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Song | 2020 | NA | 10.3760/cma.j.cn112140-20200317- 00258 | Conference abstract |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2495 | c.2495T>C | F832S | p.Phe832Ser | Activating | ADH1 | 122284449 | NA | SNV | Missense | Exon 7 | TMD | VUS | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 2501 | c.2501delC | S834fs* | p.Ser834fs* | Inactivating | FHH1 (heterozygous) | 122284455 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Ma | 2008 | 18525093 | https://www.hkmj.org/abstracts/v14n3/226.htm | Journal article |
| 2503 | c.2503G>A | A835T | p.Ala835Thr | Activating | ADH1 | 122284457 | rs2074939751 | SNV | Missense | Exon 7 | TMD | VUS | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 2504 | c.2504C>A | A835D | p.Ala835Asp | Activating | ADH1 | 122284458 | NA | SNV | Missense | Exon 7 | TMD | NA | Letz | 2014 | 25506941 | 10.1371/journal.pone.0115178 | Journal article |
| 2506 | c.2506G>A | V836I | p.Val836Ile | Activating | ADH1 | 122284460 | NA | SNV | Missense | Exon 7 | TMD | NA | Eugenin | 2016 | NA | 10.1210/endo-meetings.2016.BCHVD.10.SUN-388 | Conference abstract |
| 2506 | c.2506G>C | V836L | p.Val836Leu | Activating | ADH1 | 122284460 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2511 | c.2511G>C | E837D | p.Glu837Asp | Activating | ADH1 | 122284465 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2516 | c.2516T>C | I839T | p.Ile839Thr | Activating | ADH1 | 122284470 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 2516 | c.2516T>C | I839T | p.Ile839Thr | Activating | ADH1 | 122284470 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Nakamura | 2013 | 23966241 | 10.1210/jc.2013-1974 | Journal article |
| 2516 | c.2516T>A | I839N | p.Ile839Asn | Activating | ADH1 | 122284470 | NA | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1698776/?oq=%22NM_000388.4(CASR):c.2516T%3EA%20(p.Ile839Asn)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2516T%3EA%20(p.Ile839Asn)#id_second | Clinvar |
| 2519 | c.2519C>T | A840V | p.Ala840Val | Activating | ADH1 | 122284473 | rs1553769127 | SNV | Missense | Exon 7 | TMD | VUS | Roberts | 2019 | 31063613 | 10.1002/jbmr.3747 | Journal article |
| 2519 | c.2519C>T | A840V | p.Ala840Val | Activating | ADH1 | 122284473 | rs1553769127 | SNV | Missense | Exon 7 | TMD | VUS | Shulman | 2022 | 35350394 | 10.1210/jendso/bvac031 | Journal article |
| 2519 | c.2519C>T | A840V | p.Ala840Val | Activating | ADH1 | 122284473 | rs1553769127 | SNV | Missense | Exon 7 | TMD | VUS | Winer | 2018 | 30470382 | 10.1016/j.jpeds.2018.08.010 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Fox | 2016 | 27540249 | 10.1136/archdischild-2016-311535.66 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Gevers | 2017 | NA | Gevers EF, Buck J, Ashman N, Thakker RV, Allgrove J. Continuous subcutaneous PTH infusion in autosomal dominant hypocalcaemia. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Kinoshika | 2013 | 24297799 | 10.1210/jc.2013-3430 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Kumaran | 2009 | NA | 10.1016/j.bone.2009.04.137 | Conference abstract |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Leach | 2012 | 22798347 | 10.1210/en.2012-1449 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Letz | 2014 | 25506941 | 10.1371/journal.pone.0115178 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Nakamura | 2013 | NA | Hormone Research in Paediatrics 2013 Vol. 80 Pages 52-53 | Conference abstract |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Oprea | 2022 | NA | 10.1159/000525606 | Conference abstract |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Sanda | 2008 | 18556971 | 10.1515/jpem.2008.21.4.385 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Sastre | 2021 | 34233101 | 10.1056/NEJMc2034981 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Sato | 2002 | 12107202 | 10.1210/jcem.87.7.8639 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Vallant | 2022 | 35317006 | 10.1097/TXD.0000000000001284 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 with Bartter syndrome | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Watanabe | 2002 | 12241879 | 10.1016/S0140-6736(02)09842-2 | Journal article |
| 2528 | c.2528C>A | A843E | p.Ala843Glu | Activating | ADH1 | 122284482 | rs104893706 | SNV | Missense | Exon 7 | TMD | Pathogenic | Zhao | 1999 | 10217436 | 10.1016/s0014-5793(99)00368-3 | Journal article |
| 2530 | c.2530G>A | A844T | p.Ala844Thr | Activating | ADH1 | 122284484 | NA | SNV | Missense | Exon 7 | TMD | NA | Letz | 2010 | 20668040 | 10.1210/jc.2010-0651 | Journal article |
| 2530 | c.2530G>C | A844P | p.Ala844Pro | Activating | ADH1 | 122284484 | NA | SNV | Missense | Exon 7 | TMD | NA | Nakajima | 2009 | 19915295 | 10.2169/internalmedicine.48.2459 | Journal article |
| 2530 | c.2530G>A | A844T | p.Ala844Thr | Activating | ADH1 | 122284484 | NA | SNV | Missense | Exon 7 | TMD | NA | Raue | 2011 | 21645025 | 10.1111/j.1365-2265.2011.04142.x | Journal article |
| 2531 | c.2531C>T | A844V | p.Ala844Val | Activating | ADH1 | 122284485 | rs2074940195 | SNV | Missense | Exon 7 | TMD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/985582/?oq=%22NM_000388.4(CASR):c.2531C%3ET%20(p.Ala844Val)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2531C%3ET%20(p.Ala844Val) | Clinvar |
| 2532 | c.2532_2539delCAGCTTT | S845fs*133 | p.Ser845fs*133 | Inactivating | FHH1 (heterozygous) | 122284486 | NA | Small deletion | Frameshift | Exon 7 | TMD | NA | Bulus | 2022 | NA | 10.1159/000525606 | Conference abstract |
| 2533 | c.2533_2545del | S845fs* | p.Ser845fs* | NA | NA | 122284486 | NA | Small deletion | Frameshift | Exon 7 | TMD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1408979/?oq=%22NM_000388.4(CASR):c.2533_2545del%20(p.Ser845fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2533_2545del%20(p.Ser845fs)#id_second | Clinvar |
| 2534 | c.2534G>A | S845N | p.Ser845Asn | Activating | ADH1 | 122284488 | NA | SNV | Missense | Exon 7 | TMD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2546 | c.2546T>C | L849P | p.Leu849Pro | Inactivating | FHH1 (heterozygous) | 122284500 | rs2074940343 | SNV | Missense | Exon 7 | TMD | VUS | Frank-Raue | 2011 | 21521328 | 10.1111/j.1365-2265.2011.04059.x | Journal article |
| 2546 | c.2546T>C | L849P | p.Leu849Pro | Inactivating | FHH1 | 122284500 | rs2074940343 | SNV | Missense | Exon 7 | TMD | VUS | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 2550 | c.2550_2551insCCAG | C850fs*132 | p.Cys850fs*132 | Inactivating | FHH1 (heterozygous) | 122284504 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | D'Souza-Li | 2002 | 11889203 | 10.1210/jcem.87.3.8280 | Journal article |
| 2550 | c.2550_2551insCCAG | C850fs*132 | p.Cys850fs*132 | Inactivating | FHH1 | 122284504 | NA | Small insertion | Frameshift | Exon 7 | TMD | NA | Cole | 2009 | 19179454 | 10.1677/JME-08-0164 | Journal article |
| 2570 | c.2570T>G | I857S | p.Ile857Ser | Activating | ADH1 with Bartter syndrome (heterozygous) | 122284524 | NA | SNV | Missense | Exon 7 | TMD | NA | Chou | 2022 | 35192837 | 10.1016/j.yexcr.2022.113080 | Journal article |
| 2579 | c.2579T>G | I860S | p.Ile860Ser | Inactivating | FHH1 | 122284533 | NA | SNV | Missense | Exon 7 | TMD | NA | Mouly | 2019 | 32347971 | 10.1111/cen.14211 | Journal article |
| 2579 | c.2579T>G | I860S | p.Ile860Ser | Inactivating | FHH1 | 122284533 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2582 | c.2582T>C | L861P | p.Leu861Pro | Inactivating | FHH1 | 122284536 | NA | SNV | Missense | Exon 7 | TMD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2600 | c.2600A>T | N867I | p.Asn867Ile | Inactivating | FHH1 | 122284554 | NA | SNV | Missense | Exon 7 | ICD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2608 | c.2608G>T | E870* | p.Glu870* | Inactivating | FHH1 | 122284562 | NA | SNV | Nonsense | Exon 7 | ICD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2611 | c.2611G>T | E871* | p.Glu871* | NA | NA | 122284565 | rs1057520646 | SNV | Nonsense | Exon 7 | ICD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/379567/?oq=%22NM_000388.4(CASR):c.2611G%3ET%20(p.Glu871Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2611G%3ET%20(p.Glu871Ter)#id_first | Clinvar |
| 2618 | c.2618G>A | R873H | p.Arg873His | Inactivating | FHH1 | 122284572 | NA | SNV | Missense | Exon 7 | ICD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2618 | c.2618G>C | R873P | p.Arg873Pro | Inactivating | FHH1 | 122284572 | NA | SNV | Missense | Exon 7 | ICD | VUS | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 2627 | c.2627C>A | T876N | p.Thr876Asn | Inactivating | FHH1 | 122284581 | NA | SNV | Missense | Exon 7 | ICD | NA | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2641 | c.2641T>C | F881L | p.Phe881Leu | Inactivating | FHH1 (heterozygous) | 122284595 | rs104893704 | SNV | Missense | Exon 7 | ICD | Likely pathogenic | Carling | 2000 | 10843194 | 10.1210/jcem.85.5.6477 | Journal article |
| 2641 | c.2641T>C | F881L | p.Phe881Leu | Inactivating | FHH1 | 122284595 | rs104893704 | SNV | Missense | Exon 7 | ICD | Likely pathogenic | Lu | 2009 | 19759318 | 10.1124/jpet.109.159228 | Journal article |
| 2644 | c.2644A>T | K882* | p.Lys882* | Inactivating | FHH1 | 122284598 | rs193922437 | SNV | Nonsense | Exon 7 | ICD | Likely pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/35793/?oq=%22NM_000388.4(CASR):c.2644A%3ET%20(p.Lys882Ter)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2644A%3ET%20(p.Lys882Ter) | Clinvar |
| 2647 | c.2647G>A | V883M | p.Val883Met | Activating | ADH1 | 122284601 | NA | SNV | Missense | Exon 7 | ICD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2654 | c.2654C>A | A885D | p.Ala885Asp | Inactivating | FHH1 (heterozygous) | 122284608 | NA | SNV | Missense | Exon 7 | ICD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2656 | c.2656C>T | R886P | p.Arg886Pro | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Christensen | 2008 | 18410554 | 10.1111/j.1365-2265.2008.03259.x | Journal article |
| 2656 | c.2656C>T | R886P | p.Arg886Pro | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Christensen | 2009 | 19250271 | 10.1111/j.1365-2265.2009.03557.x | Journal article |
| 2656 | c.2656C>T | R886W | p.Arg886Trp | Inactivating | NSHPT (homozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2656 | c.2656C>T | R886W | p.Arg886Trp | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Jakobsen | 2013 | 23764372 | 10.1530/EJE-13-0224 | Journal article |
| 2656 | c.2656C>T | R886W | p.Arg886Trp | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Nissen | 2007 | 17698911 | 10.1210/jc.2007-0322 | Journal article |
| 2656 | c.2656C>T | R886P | p.Arg886Pro | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Reschke | 2013 | NA | Reschke F, Lohse J, Huebner A. A novel cytoplasmatic mutation of the calcium-sensing receptor (CASR) in a newborn resulting in familial hypocalciuric hypercalcaemia (FHH). Hormone Research in Paediatrics. 2013;80:223-4. | Conference abstract |
| 2656 | c.2656C>T | R886P | p.Arg886Pro | Inactivating | FHH1 (heterozygous) | 122284610 | rs1559969429 | SNV | Missense | Exon 7 | ICD | Pathogenic | Simonds | 2002 | 11807402 | 10.1097/00005792-200201000-00001 | Journal article |
| 2657 | c.2657G>C | R886P | p.Arg886Pro | Inactivating | FHH1 (heterozygous) | 122284611 | rs1057520791 | SNV | Missense | Exon 7 | ICD | Pathogenic | Baral | 2017 | NA | Baral S, Mamillapalli CK, Aziz SW, Jakoby MG. Calcium-sensing receptor mutation positive hypercalcemia with clinical features of primary hyperparathyroidism. Endocrine Reviews. 2017;38(3). | Conference abstract |
| 2659 | c.2659G>C | A887P | p.Ala887Pro | Inactivating | FHH1 (heterozygous) | 122284613 | NA | SNV | Missense | Exon 7 | ICD | NA | Mariathasan | 2020 | 32430905 | 10.1111/cen.14254 | Journal article |
| 2660 | c.2660C>A | A887D | p.Ala887Asp | Inactivating | FHH1 (heterozygous) | 122284614 | NA | SNV | Missense | Exon 7 | ICD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2663 | c.2663C>T | T888M | p.Thr888Met | Activating | ADH1 | 122284617 | NA | SNV | Missense | Exon 7 | ICD | NA | Lazarus | 2011 | 21135065 | 10.1530/EJE-10-0907 | Journal article |
| 2682 | c.2682delC | S895Pfs*44 | p.Ser895Profs*44 | Activating | ADH1 | 122284636 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Cisternino | 2013 | NA | Cisternino M, Andrea Bassi L, Rossetti G, Bulzomí P, Codazzi AC, Magrassi S, et al. A novel de novo activating mutation of calcium-sensing receptor (CaSR) in a patient with hypocalcemia and long QT syndrome (LQTS). Hormone Research in Paediatrics. 2013;80:225. | Conference abstract |
| 2682 | c.2682_3224del | S895_V1075del | p.Ser895_V1075del | Activating | ADH1 | 122284636 | rs1553769169 | CNV | Deletion | Exon 7 | ICD | Pathogenic | Lienhardt | 2000 | 10770217 | 10.1210/jcem.85.4.6570 | Journal article |
| 2682 | c.2682_3224del | S895_V1075del | p.Ser895_V1075del | Activating | ADH1 | 122284636 | rs1553769169 | CNV | Deletion | Exon 7 | ICD | Pathogenic | Lienhardt | 2001 | 11701698 | 10.1210/jcem.86.11.8016 | Journal article |
| 2682 | c.2682delC | S895Pfs*44 | p.Ser895Profs*44 | Activating | ADH1 | 122284636 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Maruca | 2017 | 27561204 | 10.1016/j.mce.2016.08.032 | Journal article |
| 2703 | c.2703_2710delCCTTGGAG | S901fs*77 | p.Gln901fs*77 | Activating | ADH1 | 122284657 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Obermannova | 2016 | 26764418 | 10.1530/EJE-15-1216 | Journal article |
| 2730 | c.2730delC | P910fs*29 | p.Pro910fs*29 | Activating | ADH1 | 122284684 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Hannan | 2012 | 22422767 | 10.1093/hmg/dds105 | Journal article |
| 2776 | c.2776C>T | Q926* | p.Gln926* | Inactivating | FHH1 | 122284730 | NA | SNV | Missense | Exon 7 | ICD | VUS | Nissen | 2019 | 31433865 | 10.1111/cen.14078 | Journal article |
| 2777 | c.2777A>G | Q926R | p.Gln926Arg | Inactivating | FHH1 (heterozygous) | 122284731 | rs200263975 | SNV | Missense | Exon 7 | ICD | Conflicting interpretations of pathogenicity | Frank-Raue | 2011 | 21521328 | 10.1111/j.1365-2265.2011.04059.x | Journal article |
| 2777 | c.2777A>G | Q926R | p.Gln926Arg | Inactivating | FHH1 | 122284731 | rs200263975 | SNV | Missense | Exon 7 | ICD | Conflicting interpretations of pathogenicity | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 2800 | c.2800C>T | Q934* | p.Gln934* | Activating | ADH1 | 122284754 | NA | SNV | Nonsense | Exon 7 | ICD | NA | García-Castaño | 2018 | 30407919 | 10.1530/EJE-18-0129 | Journal article |
| 2901 | c.2901delC | L968fs*10 | p.Leu968fs*10 | Activating | ADH1 | 122284855 | NA | SNV | Frameshift | Exon 7 | ICD | NA | Nissen | 2012 | 22192860 | 10.1016/j.cca.2011.12.004 | Journal article |
| 2901 | c.2901del | F968fs* | p.Phe968fs* | NA | NA | 122284855 | NA | Small deletion | Frameshift | Exon 7 | ICD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/1697473/?oq=%22NM_000388.4(CASR):c.2901del%20(p.Phe968fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.2901del%20(p.Phe968fs)#id_second | Clinvar |
| 2915 | c.2915C>T | T972M | p.Thr972Met | Inactivating | FHH1 (heterozygous) | 122284869 | rs200620134 | SNV | Missense | Exon 7 | ICD | Conflicting interpretations of pathogenicity | Mastromatteo | 2014 | 25292184 | 10.1186/1472-6823-14-81 | Journal article |
| 3010 | c.3010del | S1014fs* | p.Ser1014fs* | NA | NA | 122284964 | rs1131691620 | Small deletion | Frameshift | Exon 7 | ICD | Pathogenic | Clinvar | NA | NA | https://www.ncbi.nlm.nih.gov/clinvar/variation/429831/?oq=%22NM_000388.4(CASR):c.3010del%20(p.Ser1004fs)%22%5Bvarname%5D&m=NM_000388.4(CASR):c.3010del%20(p.Ser1004fs)#id_second | Clinvar |
| 3013 | c.3013G>A | D1005N | p.Asp1005Asn | Inactivating | FHH1 (heterozygous) | 122284967 | rs201990892 | SNV | Missense | Exon 7 | ICD | VUS | Frank-Raue | 2011 | 21521328 | 10.1111/j.1365-2265.2011.04059.x | Journal article |
| 3013 | c.3013G>A | D1005N | p.Asp1005Asn | Inactivating | FHH1 | 122284967 | rs201990892 | SNV | Missense | Exon 7 | ICD | VUS | Rus | 2008 | 18796518 | 10.1210/jc.2008-1076 | Journal article |
| 3104 | c.3104delC | P1025Lfs*21 | p.Pro1025Leufs*21 | Inactivating | FHH1 | 122285058 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Vargas-Poussou | 2016 | 26963950 | 10.1210/jc.2015-3442 | Journal article |
| 3163 | c.3163_3170delinsTTACTGGGGA | V1045fs* | p.Val1045fs* | Inactivating | FHH1 (heterozygous) | 122285117 | NA | SNV | Frameshift | Exon 7 | ICD | NA | Mijares Zamuner | 2013 | 22789152 | 10.1016/j.endonu.2012.04.006 | Journal article |
| 3193 | c.3193delA | S1065Vfs* | p.Ser1065Vfs* | Inactivating | FHH1 (heterozygous) | 122285147 | NA | Small deletion | Frameshift | Exon 7 | ICD | NA | Yabuta | 2009 | 19423460 | 10.1016/S1015-9584(09)60022-1 | Journal article |
| 3235 | c.3235T>C | *1079Qext*8 | p.1079Glnext8 | Inactivating | FHH1 (heterozygous) | 122285189 | NA | SNV | Loss of stop codon | Exon 7 | ICD | NA | Guarnieri | 2010 | 20164288 | 10.1210/jc.2008-2430 | Journal article |
| 3235 | c.3235T>C | *1079Qext*8 | p.1079Glnext8 | Inactivating | FHH1 (heterozygous) | 122285189 | NA | SNV | Loss of stop codon | Exon 7 | ICD | NA | Tellam | 2020 | 32698162 | 10.1530/EDM-20-0004 | Journal article |
| NA | NA | V268fs*6 | p.Val268fs*6 | Inactivating | FHH1 (heterozygous) | NA | NA | Small deletion | Frameshift | Exon 3 | ECD | NA | Simonds | 2002 | 11807402 | 10.1097/00005792-200201000-00001 | Journal article |
| NA | NA | T876insAlufs-1X911 | p.Thr876insAlufs-1X911 | Inactivating | FHH1 (heterozygous) | NA | NA | Small insertion | Frameshift | Exon 7 | ICD | NA | Janicic | 1995 | 7717399 | NA | Journal article |
| NA | NA | T876insAlufs-1X911 | p.Thr876insAlufs-1X911 | Inactivating | NSHPT (homozygous) | NA | NA | Small insertion | Frameshift | Exon 7 | ICD | NA | Janicic | 1995 | 7717399 | NA | Journal article |
Note: Amino acid positions in the database are referenced according to the P41180-1 isoform, which consists of 1,078 amino acids. Genomic coordinates in the database are referenced according to the GRCh38 genome assembly.
Amino Acid Snake Diagram
Summary Statistics
- Number of Variants
- Variant Count by Exon
- Variant Count by Structural Domain
- Frequently Reported Variants
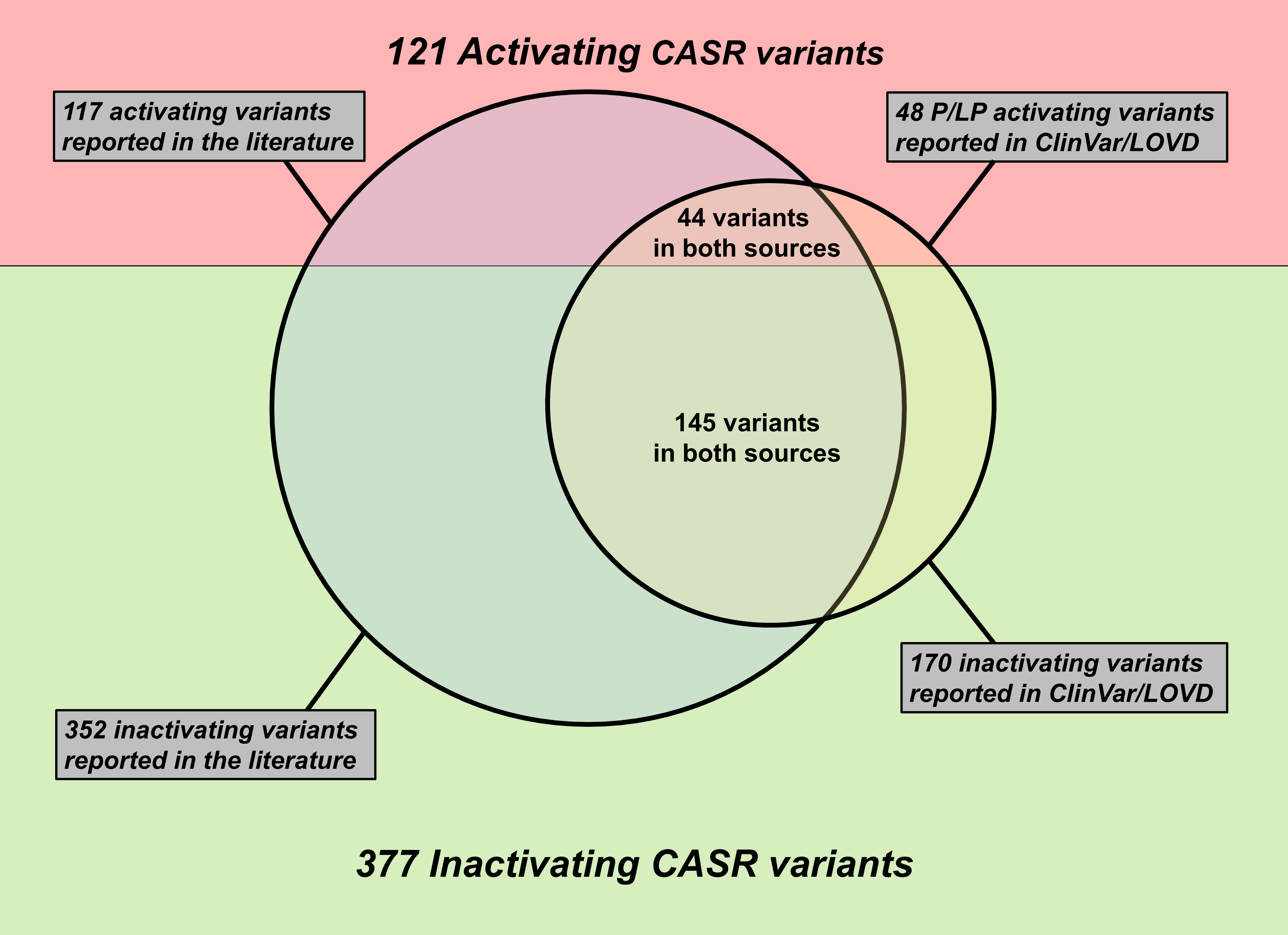
We identified a total of 498 variants, of which 121 (24.3%) were associated with ADH1 and 377 (75.7%) with FHH1. Most variants included in our database were identified from the literature (117 activating and 352 inactivating variants), and the majority of these were not documented in ClinVar/LOVD (73/117, 62.4% activating variants; 207/352, 58.8% inactivating variants). A total of 44 activating and 145 inactivating variants were identified from both the literature and ClinVar/LOVD. Only 25 inactivating and 4 activating variants were reported as P/LP in ClinVar/LOVD but not found in the literature. Furthermore, 52 P/LP variants were noted only in ClinVar/LOVD without an associated phenotype.
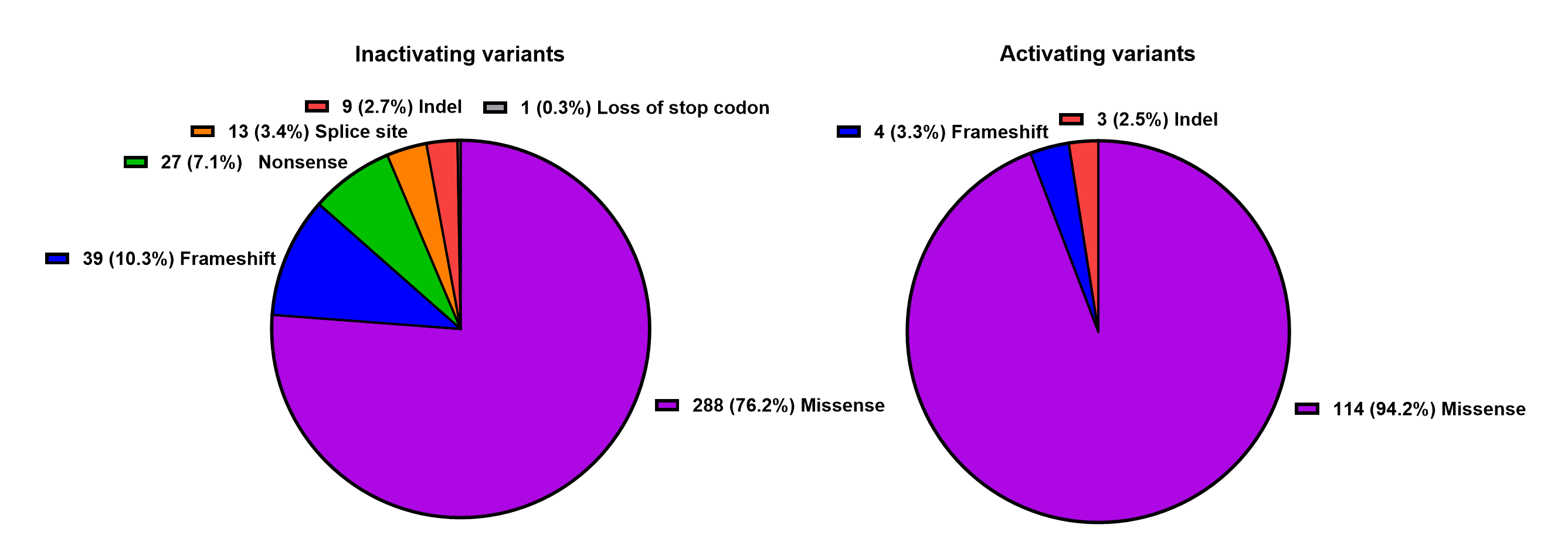
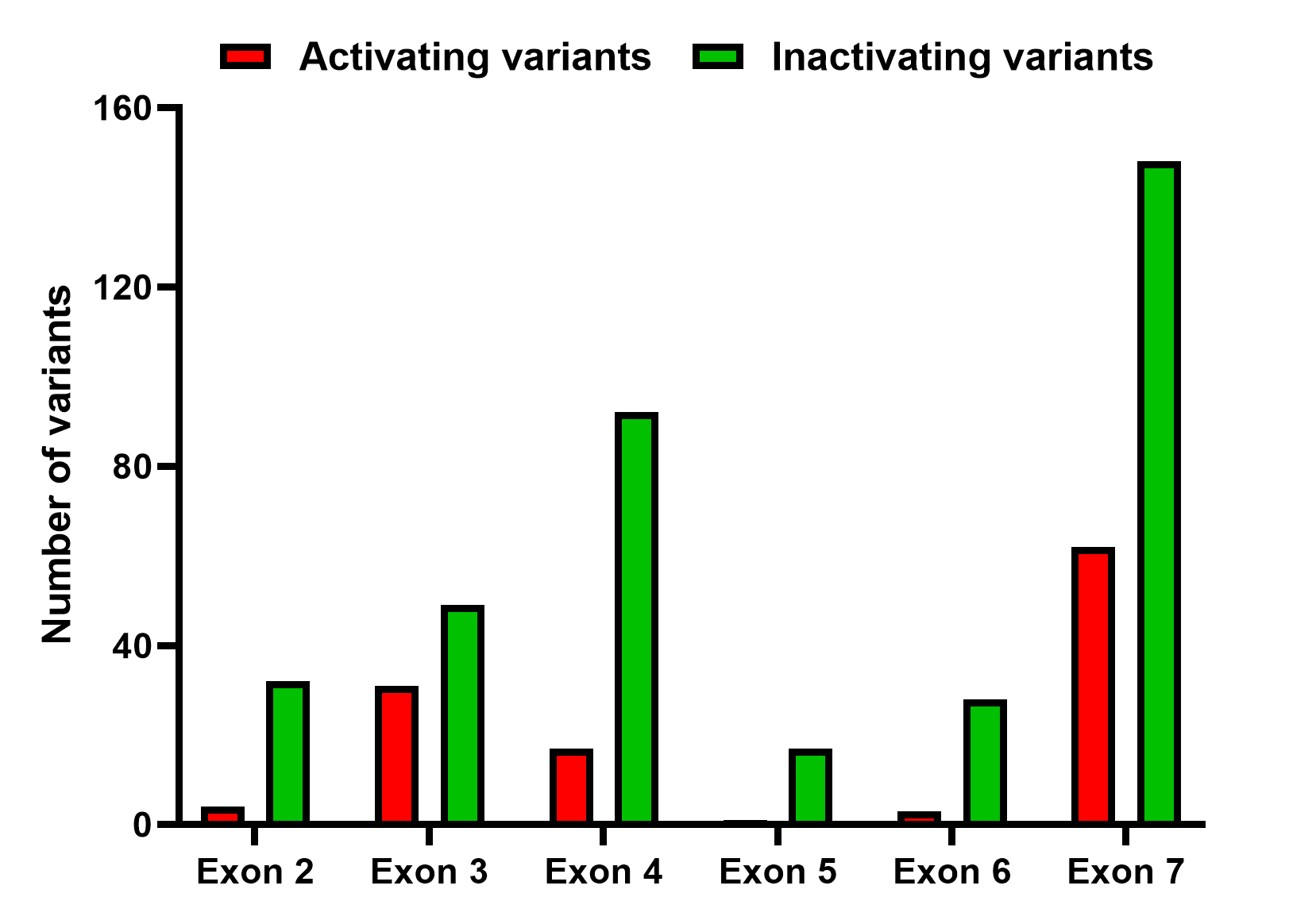
Activating variants: Exon 2: 4; Exon 3: 31; Exon 4: 17; Exon 5: 1; Exon 6: 3; Exon 7: 62
Inactivating variants: Exon 2: 32; Exon 3: 49; Exon 4: 92; Exon 5: 17; Exon 6: 28; Exon 7: 149
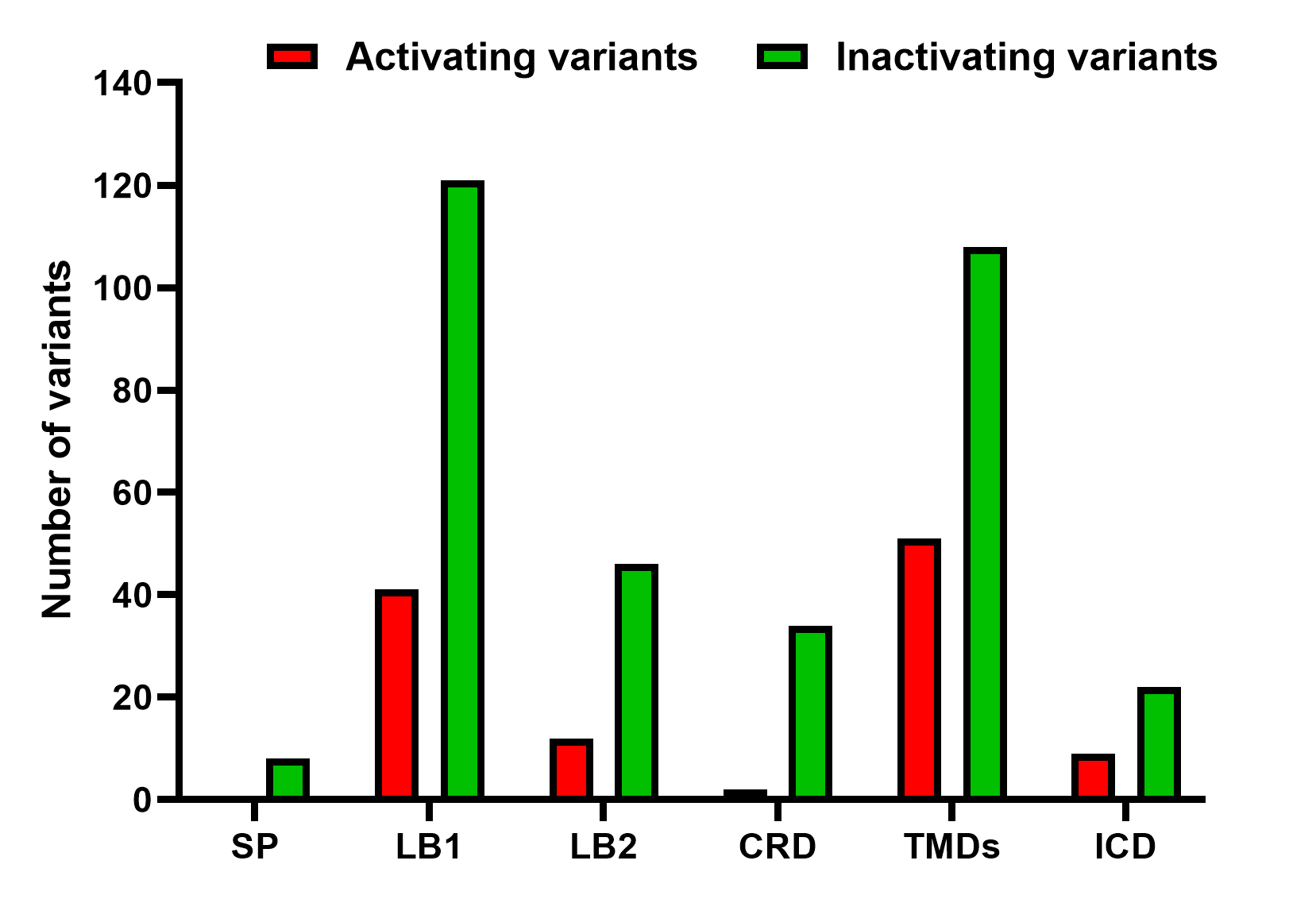
Activating variants: SP: 0; LB1: 41; LB2: 12; CRD: 4; TMDs: 51; ICD: 9
Inactivating variants: SP: 8; LB1: 121; LB2: 46; CRD: 34; TMDs: 108; ICD: 22
Abbreviations: SP: Signal Peptide (Met1 – Ala19) ; LB1: Lobe 1 (Tyr20 – Asn189 and Lys323 – Gly487); LB2: Lobe 2 (Asp190 – Leu322 and Asn488 – Asn541); CRD: Cysteine-rich Domain (Cys542 – Ile599); TMDs: Transmembrane Domains (Ala600 – Ile859); ICD: Intracellular Domain (Ile860 – Ser1078)